Содержание
- 2. Watson and Crick 1953 article in Nature
- 3. Double helix structure of DNA “It has not escaped our notice that the specific pairing we
- 4. Directionality of DNA You need to number the carbons! it matters! OH CH2 O 4′ 5′
- 5. The DNA backbone Putting the DNA backbone together refer to the 3′ and 5′ ends of
- 6. Anti-parallel strands Nucleotides in DNA backbone are bonded from phosphate to sugar between 3′ & 5′
- 7. Bonding in DNA ….strong or weak bonds? How do the bonds fit the mechanism for copying
- 8. Base pairing in DNA Purines adenine (A) guanine (G) Pyrimidines thymine (T) cytosine (C) Pairing A
- 9. Copying DNA Replication of DNA base pairing allows each strand to serve as a template for
- 10. Replication: 1st step Unwind DNA helicase enzyme unwinds part of DNA helix stabilized by single-stranded binding
- 11. DNA Polymerase III Replication: 2nd step But… We’re missing something! What? Where’s the ENERGY for the
- 12. energy ATP GTP TTP CTP Energy of Replication Where does energy for bonding usually come from?
- 13. Limits of DNA polymerase III can only build onto 3′ end of an existing DNA strand
- 14. Replication fork / Replication bubble leading strand lagging strand leading strand lagging strand leading strand lagging
- 15. RNA primer built by primase serves as starter sequence for DNA polymerase III Limits of DNA
- 16. DNA polymerase I removes sections of RNA primer and replaces with DNA nucleotides But DNA polymerase
- 17. Loss of bases at 5′ ends in every replication chromosomes get shorter with each replication limit
- 18. Repeating, non-coding sequences at the end of chromosomes = protective cap limit to ~50 cell divisions
- 19. Replication fork 3’ 5’ 3’ 5’ 5’ 3’ 3’ 5’ helicase SSB = single-stranded binding proteins
- 21. Скачать презентацию
 Дослідження. Пророщування насіння
Дослідження. Пророщування насіння ПРЕЗЕНТАЦИЯ ДЛЯ ИНТЕРАКТИВНОЙ ДОСКИ. Тип Кольчатые черви. 7 класс (В трёх частях)
ПРЕЗЕНТАЦИЯ ДЛЯ ИНТЕРАКТИВНОЙ ДОСКИ. Тип Кольчатые черви. 7 класс (В трёх частях) Пищевые продукты, питательные вещества и их превращения в организме
Пищевые продукты, питательные вещества и их превращения в организме Лишайники (лихенизированные грибы)
Лишайники (лихенизированные грибы) Внутренняя среда организма. Кровь
Внутренняя среда организма. Кровь Витамины и их роль в организме человека
Витамины и их роль в организме человека Морфофункциональная характеристика скелета и аппарата движения туловища
Морфофункциональная характеристика скелета и аппарата движения туловища Класс Млекопитающие, или Звери
Класс Млекопитающие, или Звери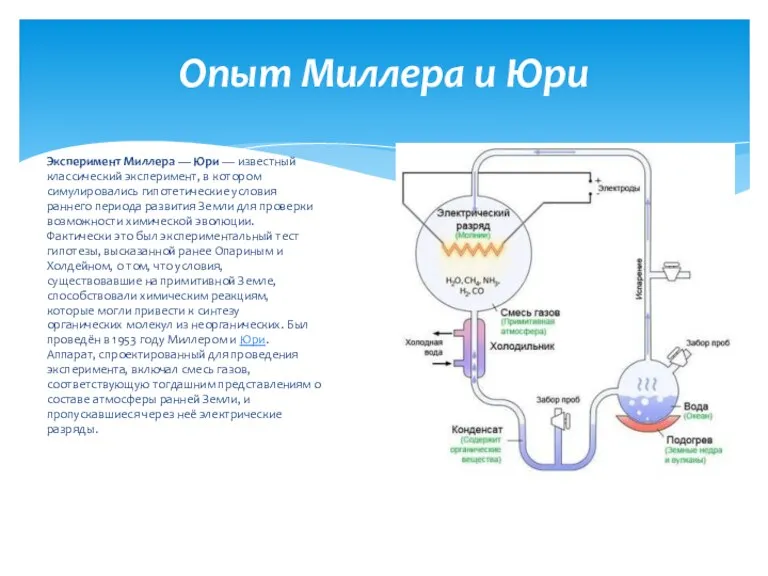 Опыт Миллера и Юри
Опыт Миллера и Юри Animals of New Zealand
Animals of New Zealand Личностно-ориентированное обучение на уроках биологии.
Личностно-ориентированное обучение на уроках биологии. Кедр європейський
Кедр європейський Обитатели пресного водоема
Обитатели пресного водоема Сезонные явления в жизни птиц
Сезонные явления в жизни птиц Сучасні критерії виду
Сучасні критерії виду Анатомо-физиологические основы голосообразования. Значение дыхания и голосообразования
Анатомо-физиологические основы голосообразования. Значение дыхания и голосообразования Витамины
Витамины Основные компоненты биотехнологической системы. Схема типового биотехнологического процесса
Основные компоненты биотехнологической системы. Схема типового биотехнологического процесса Внутренняя среда. Значение крови и ее состав. 8 класс
Внутренняя среда. Значение крови и ее состав. 8 класс Биотические отношения
Биотические отношения Современные методы исследования органа зрения
Современные методы исследования органа зрения Немного о собаках
Немного о собаках Органические вещества. Биополимеры. Углеводы
Органические вещества. Биополимеры. Углеводы Дигибридное скрещивание. Третий закон Г. Менделя
Дигибридное скрещивание. Третий закон Г. Менделя Класс Двудольные. Семейство Крестоцветные
Класс Двудольные. Семейство Крестоцветные Проверочная работа по теме Органоиды клетки.
Проверочная работа по теме Органоиды клетки. Многообразие млекопитающих. Первозвери и настоящие звери
Многообразие млекопитающих. Первозвери и настоящие звери Ас қорыту жүйесі
Ас қорыту жүйесі