Содержание
- 2. Content PHYSIOLOGICAL STUDIES 1. Nutritional studies 2. Growth studies II. GENETIC SYSTEMS 1. Genetic methods 2.
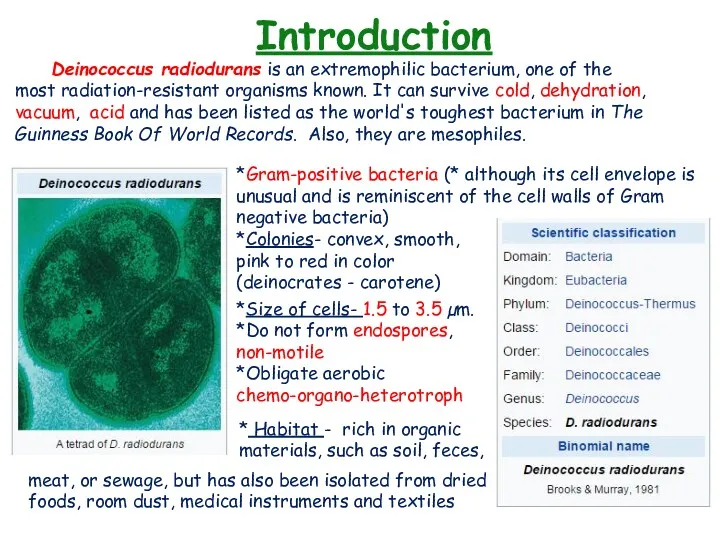
- 3. Introduction Deinococcus radiodurans is an extremophilic bacterium, one of the most radiation-resistant organisms known. It can
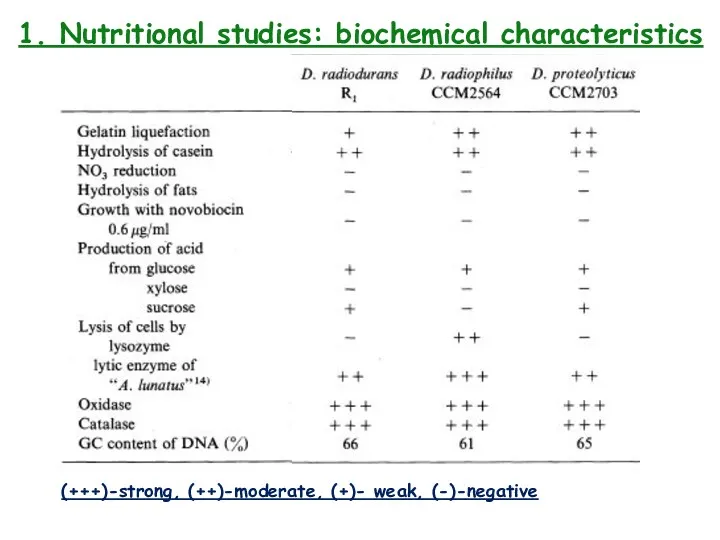
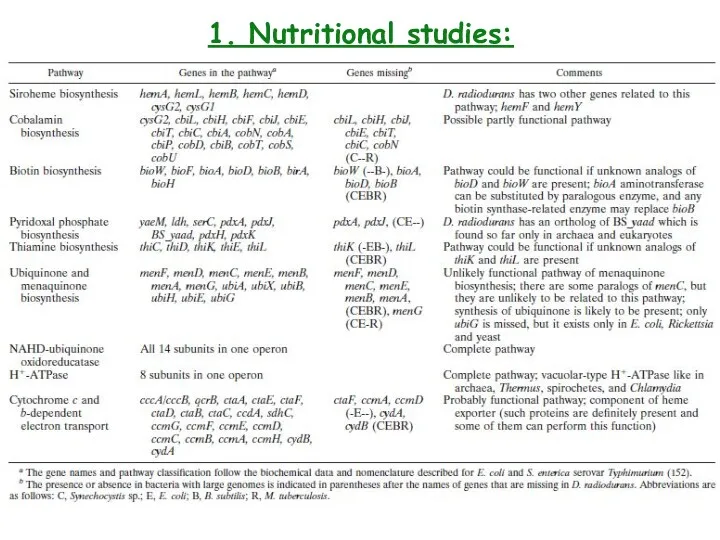
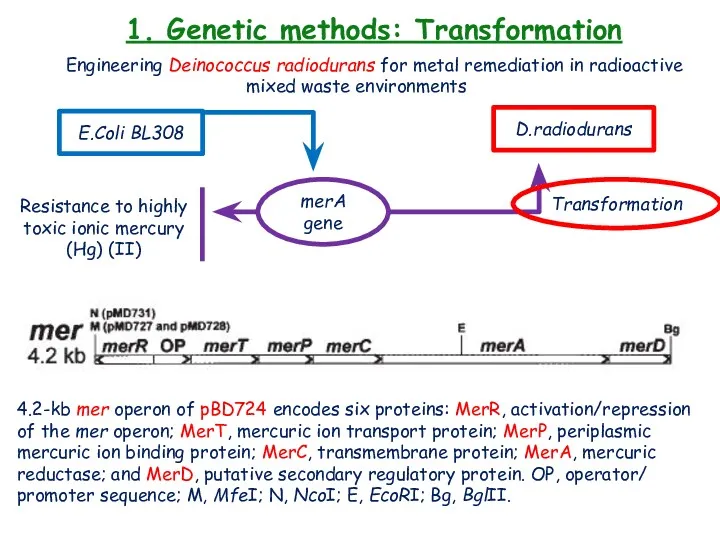
- 4. 1. Nutritional studies: biochemical characteristics (+++)-strong, (++)-moderate, (+)- weak, (-)-negative
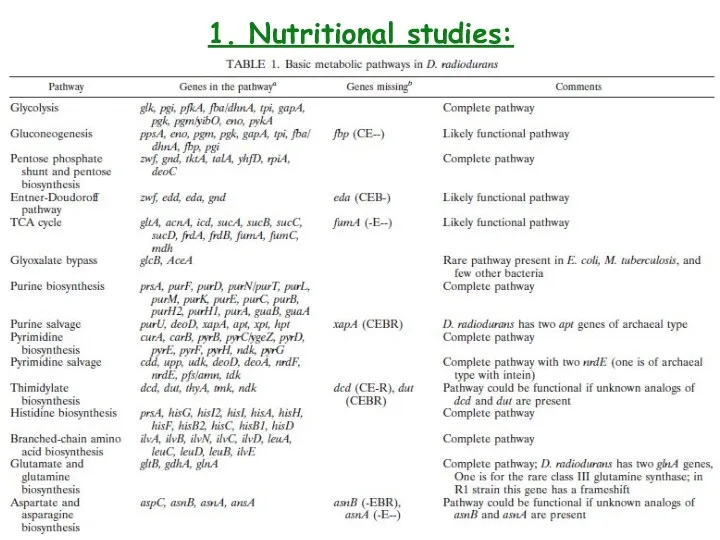
- 5. 1. Nutritional studies:
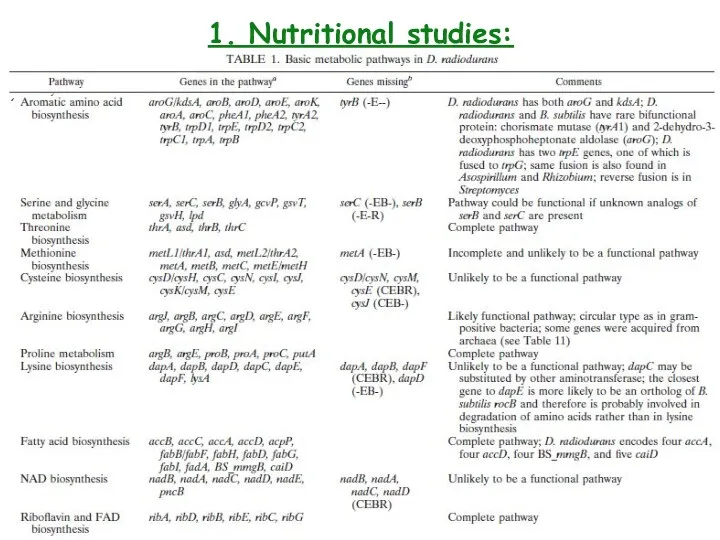
- 6. 1. Nutritional studies:
- 7. 1. Nutritional studies:
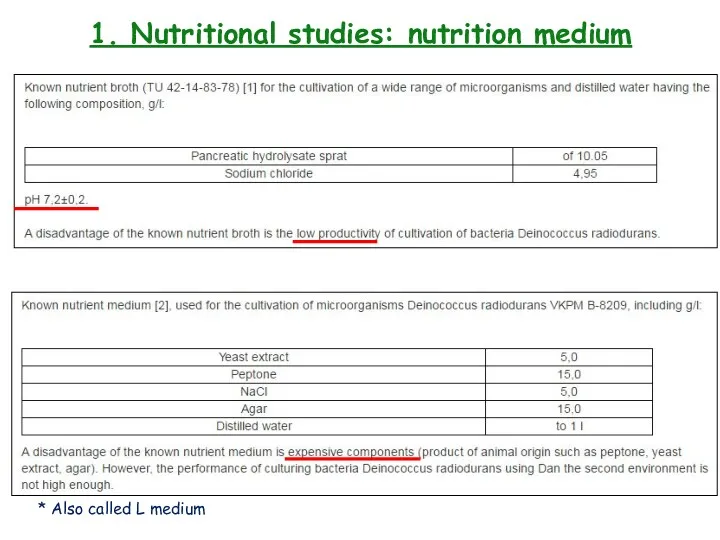
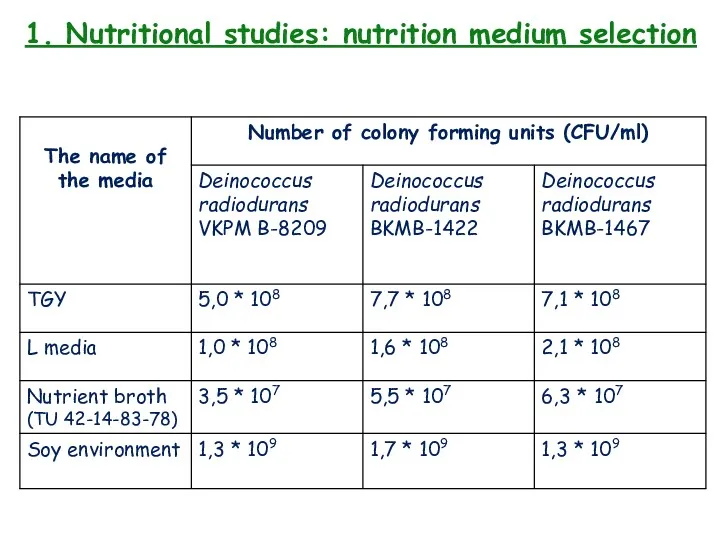
- 8. 1. Nutritional studies: nutritional medium
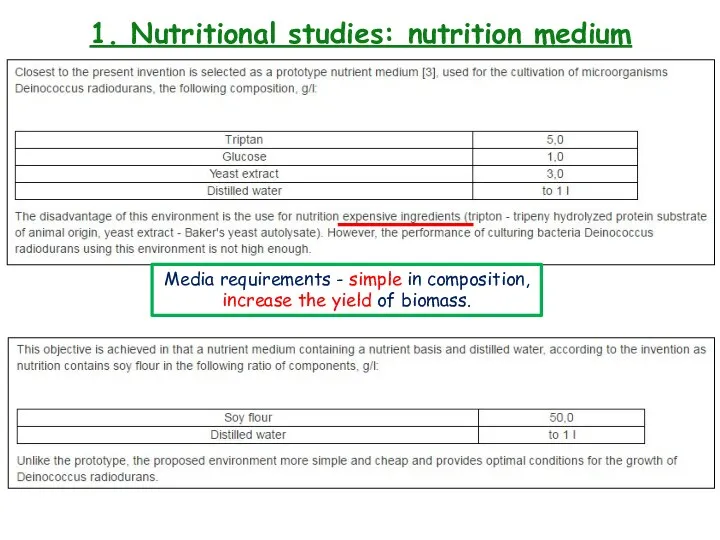
- 9. 1. Nutritional studies: nutritional medium
- 10. 1. Nutritional studies: nutrition medium * Also called L medium
- 11. 1. Nutritional studies: nutrition medium Media requirements - simple in composition, increase the yield of biomass.
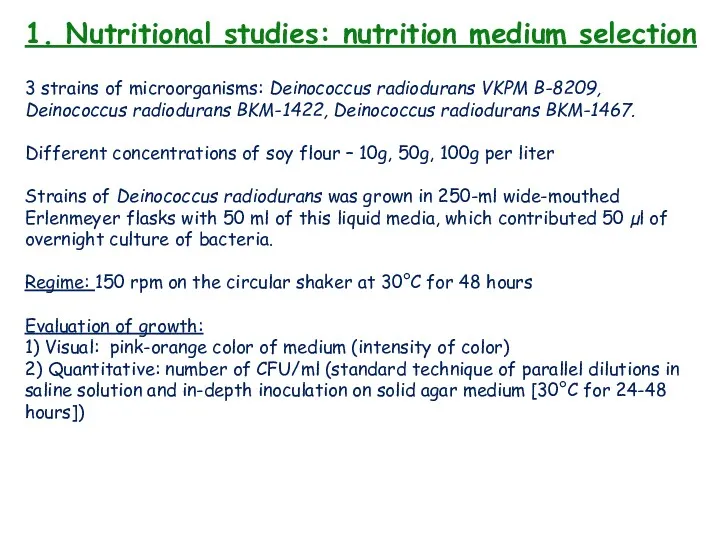
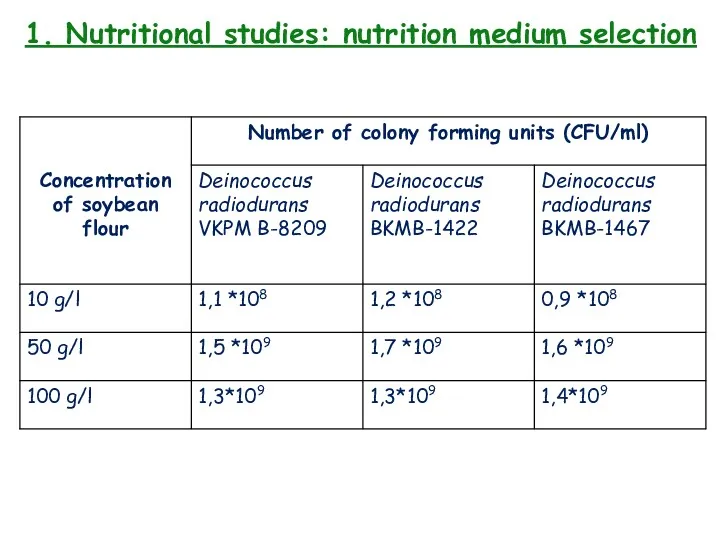
- 12. 1. Nutritional studies: nutrition medium selection 3 strains of microorganisms: Deinococcus radiodurans VKPM B-8209, Deinococcus radiodurans
- 13. 1. Nutritional studies: nutrition medium selection
- 14. 1. Nutritional studies: nutrition medium selection
- 15. Cultivation -> Solid state/submerged Bioreactor design ->Airlift bioreactor Fermentation mode -> Batch, Continuous, Semi-continuous Culture monitoring
- 16. 2. Genomic approaches The genome of D. radiodurans consists of four major parts. The complete sequence
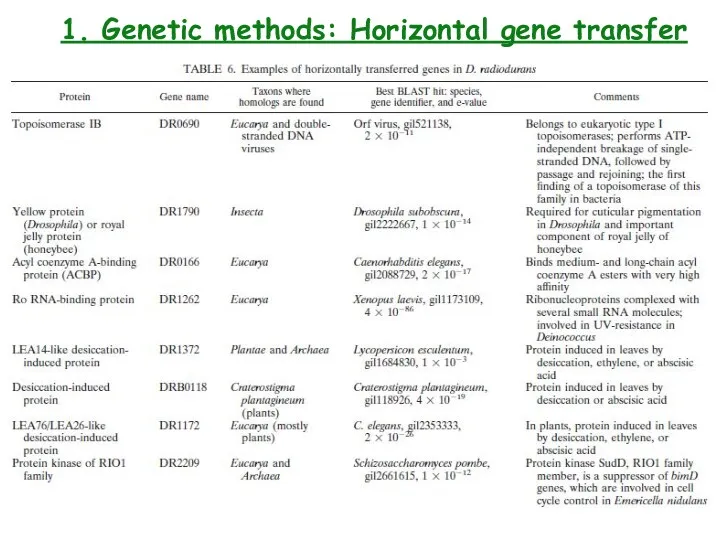
- 17. 1. Genetic methods: Horizontal gene transfer
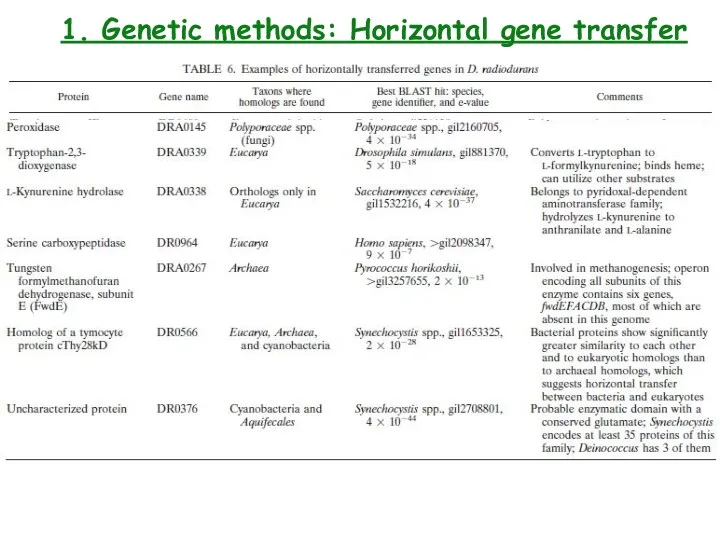
- 18. 1. Genetic methods: Horizontal gene transfer
- 19. 1. Genetic methods: Transformation Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments E.Coli
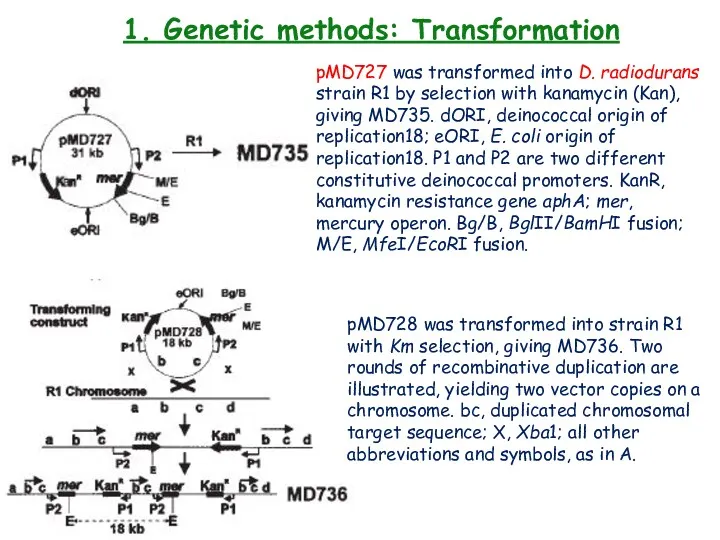
- 20. 1. Genetic methods: Transformation pMD727 was transformed into D. radiodurans strain R1 by selection with kanamycin
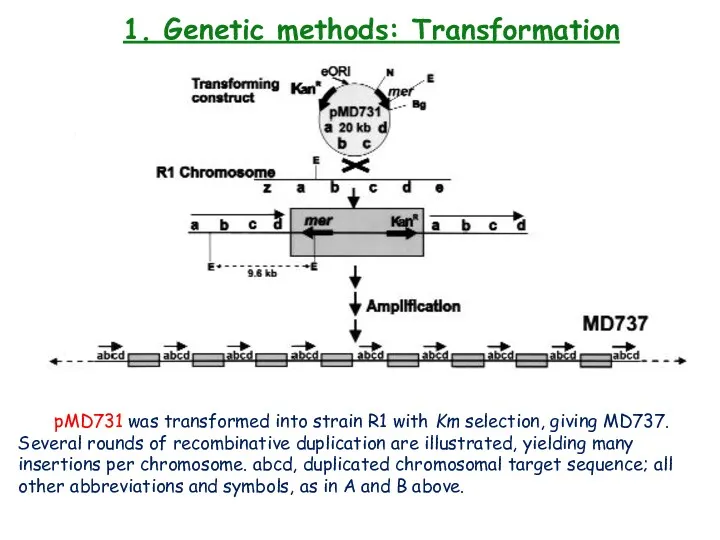
- 21. 1. Genetic methods: Transformation pMD731 was transformed into strain R1 with Km selection, giving MD737. Several
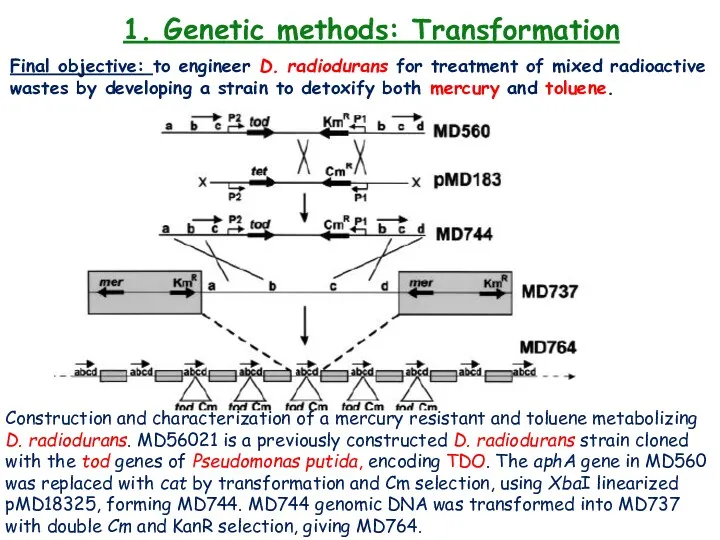
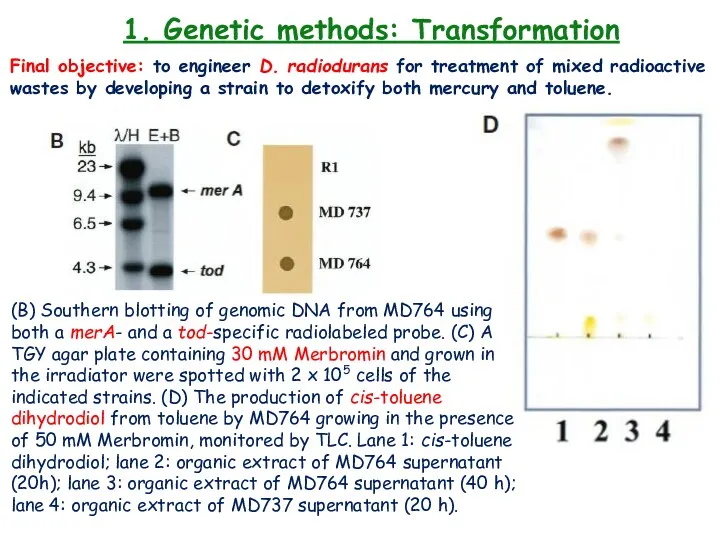
- 22. 1. Genetic methods: Transformation Final objective: to engineer D. radiodurans for treatment of mixed radioactive wastes
- 23. 1. Genetic methods: Transformation Final objective: to engineer D. radiodurans for treatment of mixed radioactive wastes
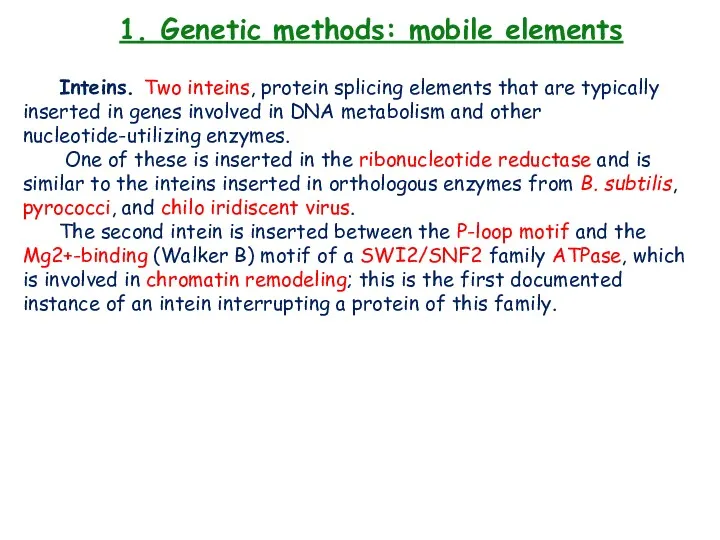
- 24. 1. Genetic methods: mobile elements Inteins. Two inteins, protein splicing elements that are typically inserted in
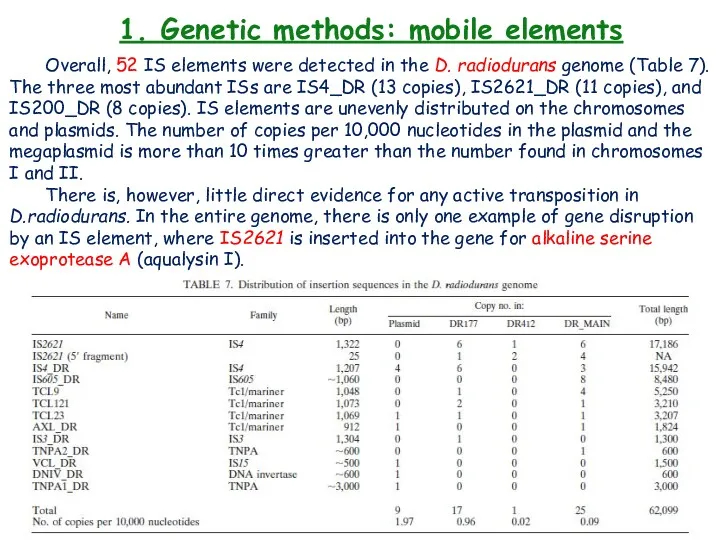
- 25. 1. Genetic methods: mobile elements Overall, 52 IS elements were detected in the D. radiodurans genome
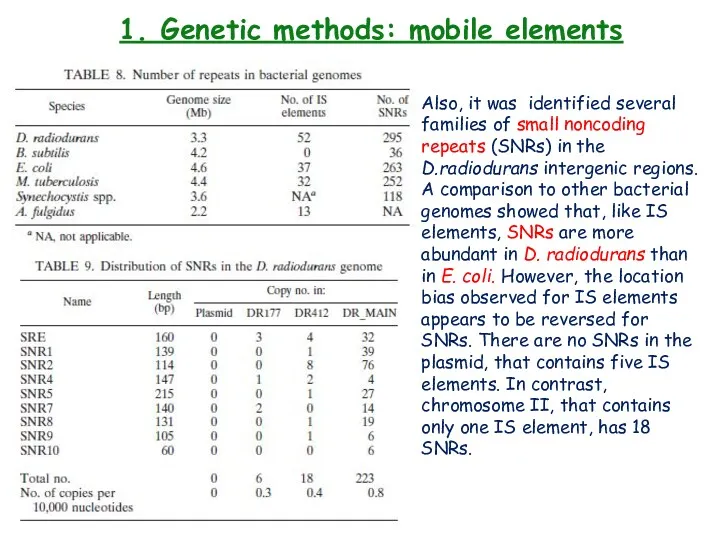
- 26. 1. Genetic methods: mobile elements Also, it was identified several families of small noncoding repeats (SNRs)
- 27. 1. Genetic methods: Gene inactivation Inactivation of Proteins Presumed to Be Involved in the Desiccation Tolerance
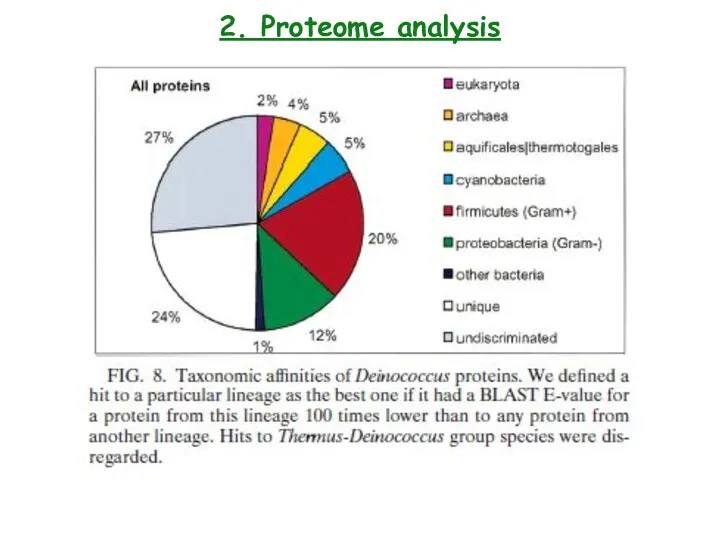
- 28. 2. Proteome analysis
- 29. 2. Proteome analysis Deinococcus radiodurans contains two thioredoxins (Trx and Trx1) and a single thioredoxin reductase
- 30. 2. Proteome analysis 3. E.coli was cultures on LB agar plates with kanamycin, Then single colonies
- 31. Reference Hitoshi Ito, Hiroshi Watanabe, Masaaki. Isolation and Identification of Radiation-resistant Cocci Belonging to the Genus
- 32. Reference John R. Battista, Mie-Jung Park, and Andrew E. McLemore. Inactivation of Two Homologues of Proteins
- 34. Скачать презентацию







 Conditional Sentences
Conditional Sentences The Verb: Mood and Modality
The Verb: Mood and Modality Peculiarities of Official Texts Translation
Peculiarities of Official Texts Translation Nouns. Техники запоминания слов This-these That-those. Articles. lesson 3
Nouns. Техники запоминания слов This-these That-those. Articles. lesson 3 Игра пред / посленовогодняя по английскому языку для учащихся 3 класса Отгадай-ка
Игра пред / посленовогодняя по английскому языку для учащихся 3 класса Отгадай-ка Past simple
Past simple Theories and perspectives in sociology
Theories and perspectives in sociology Famous people of New Zealand
Famous people of New Zealand Мастер класс. Своя игра
Мастер класс. Своя игра Spotlight 3. Module 7 (Unit 13). A Day Off
Spotlight 3. Module 7 (Unit 13). A Day Off Cultural Manners
Cultural Manners Ecological problems. What you can do to protect our planet
Ecological problems. What you can do to protect our planet What do you like to wear?
What do you like to wear? Statement of purpose
Statement of purpose Places of interests in Syktyvkar
Places of interests in Syktyvkar Happy birthday!
Happy birthday! England is the largest and major part of the United Kingdom
England is the largest and major part of the United Kingdom Merry Christmas
Merry Christmas Косвенная речь. Indirect speech
Косвенная речь. Indirect speech Superstitions in Britain
Superstitions in Britain Добро пожаловать на берега Адриатического моря. Специфика лагеря Only English Land
Добро пожаловать на берега Адриатического моря. Специфика лагеря Only English Land My profession
My profession насекомые
насекомые Present Continuous Tense. Usage
Present Continuous Tense. Usage Young Heroes of the War 1941-1945
Young Heroes of the War 1941-1945 Christmas in Britain
Christmas in Britain Household chores
Household chores Entertainment and Media
Entertainment and Media