Содержание
- 2. General pipeline Raw reads
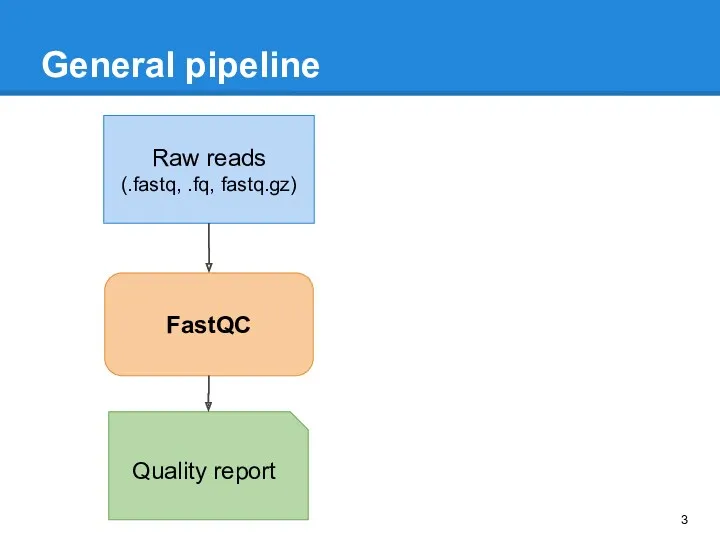
- 3. General pipeline Raw reads (.fastq, .fq, fastq.gz) FastQC Quality report
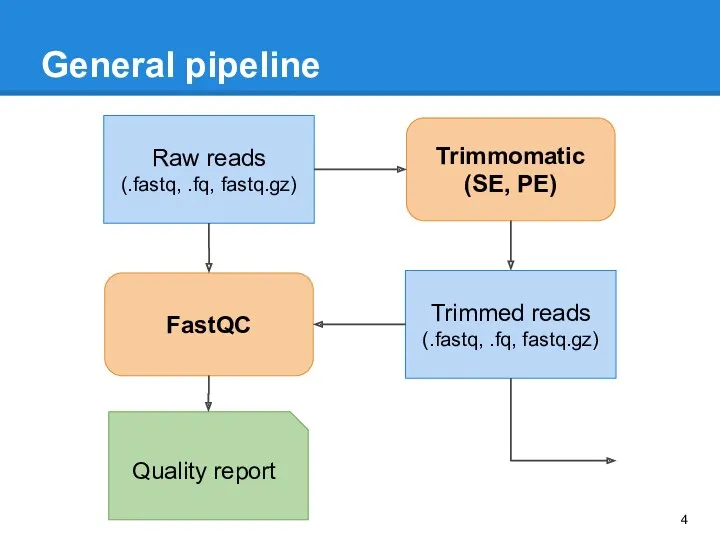
- 4. General pipeline Raw reads (.fastq, .fq, fastq.gz) FastQC Trimmomatic (SE, PE) Trimmed reads (.fastq, .fq, fastq.gz)
- 5. General pipeline Trimmed reads (.fastq, .fq, fastq.gz)
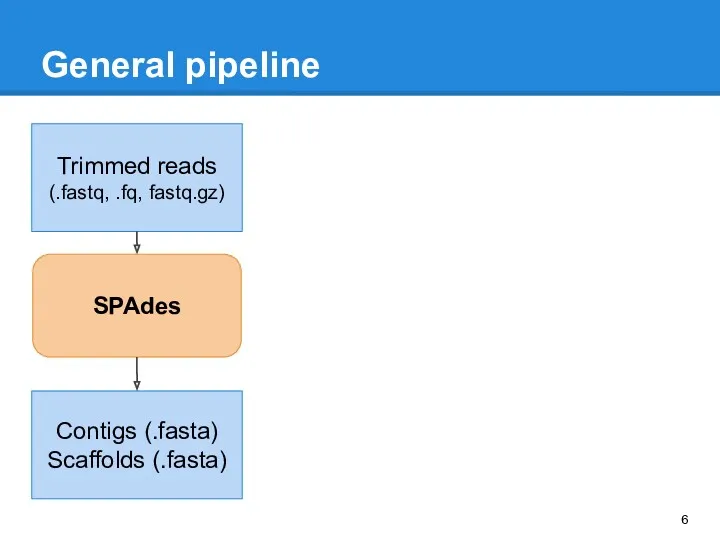
- 6. General pipeline Trimmed reads (.fastq, .fq, fastq.gz) SPAdes Contigs (.fasta) Scaffolds (.fasta)
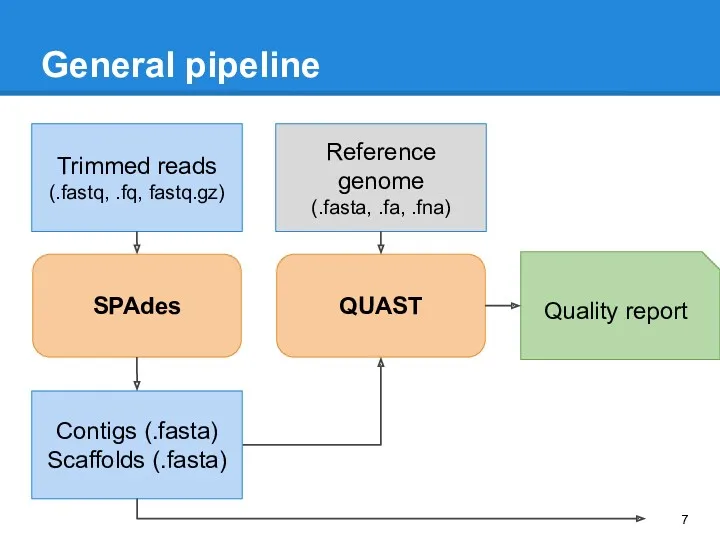
- 7. General pipeline QUAST Trimmed reads (.fastq, .fq, fastq.gz) Quality report SPAdes Contigs (.fasta) Scaffolds (.fasta) Reference
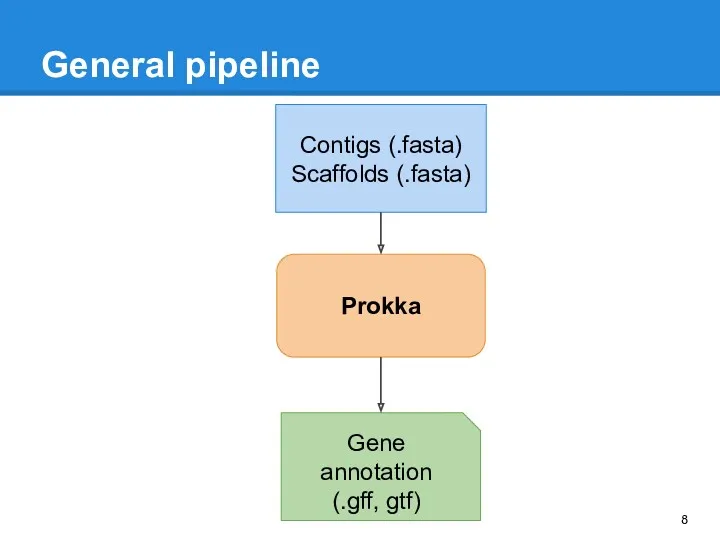
- 8. General pipeline Prokka Gene annotation (.gff, gtf) Contigs (.fasta) Scaffolds (.fasta)
- 9. Genome Annotation Questions What is the order are the genes and does this have any significance?
- 10. After completing the human genome we faced 3 Gigabytes of this: Genome sequence does not give
- 11. Not immediately apparent where the genes are…
- 12. Genomic Features Protein coding genes. In long open reading frames ORFs interrupted by introns in eukaryotes
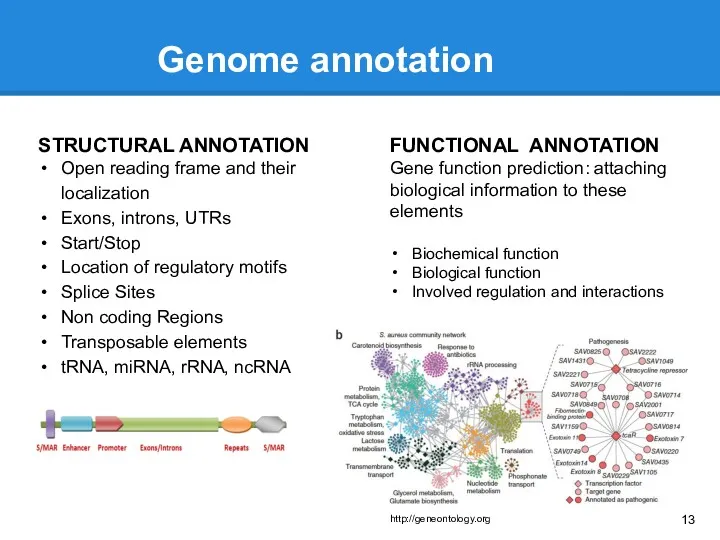
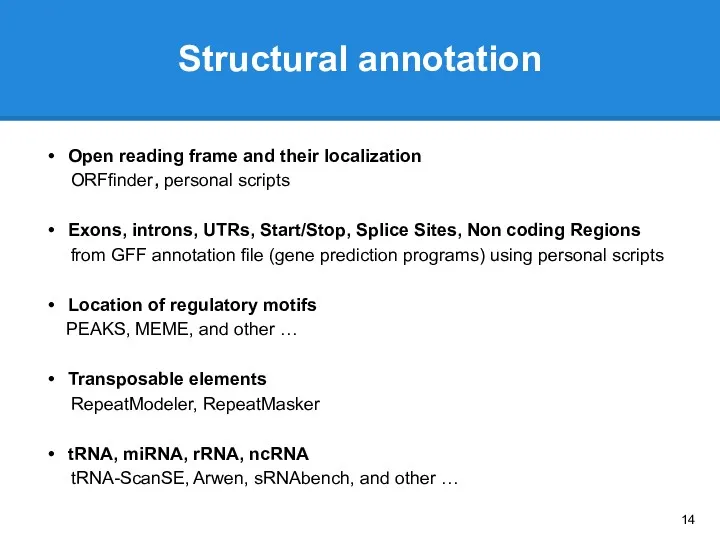
- 13. Genome annotation STRUCTURAL ANNOTATION Open reading frame and their localization Exons, introns, UTRs Start/Stop Location of
- 14. Structural annotation Open reading frame and their localization ORFfinder, personal scripts Exons, introns, UTRs, Start/Stop, Splice
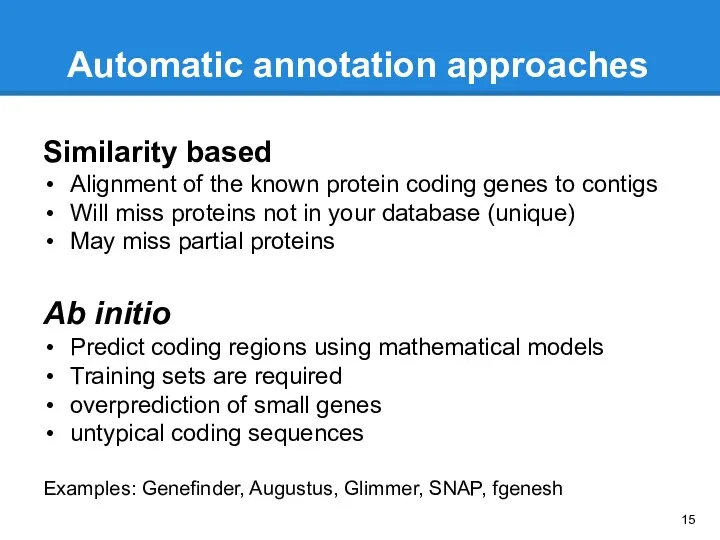
- 15. Similarity based Alignment of the known protein coding genes to contigs Will miss proteins not in
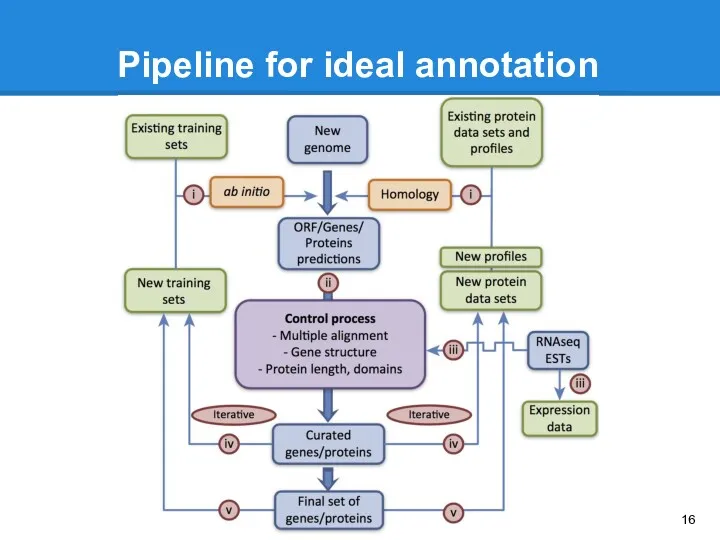
- 16. Pipeline for ideal annotation
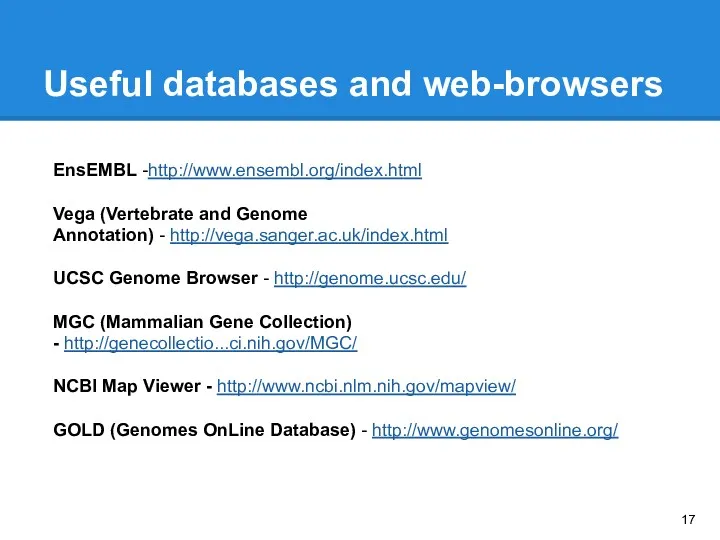
- 17. Useful databases and web-browsers EnsEMBL -http://www.ensembl.org/index.html Vega (Vertebrate and Genome Annotation) - http://vega.sanger.ac.uk/index.html UCSC Genome Browser
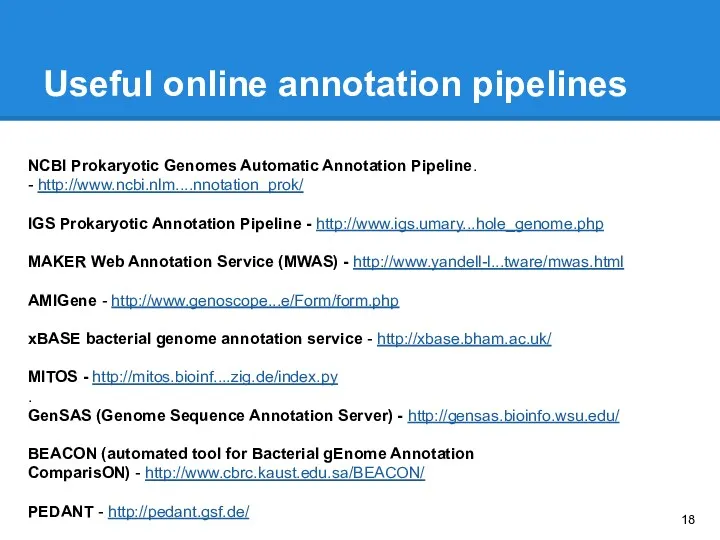
- 18. Useful online annotation pipelines NCBI Prokaryotic Genomes Automatic Annotation Pipeline. - http://www.ncbi.nlm....nnotation_prok/ IGS Prokaryotic Annotation Pipeline
- 19. Bacterial genome annotation
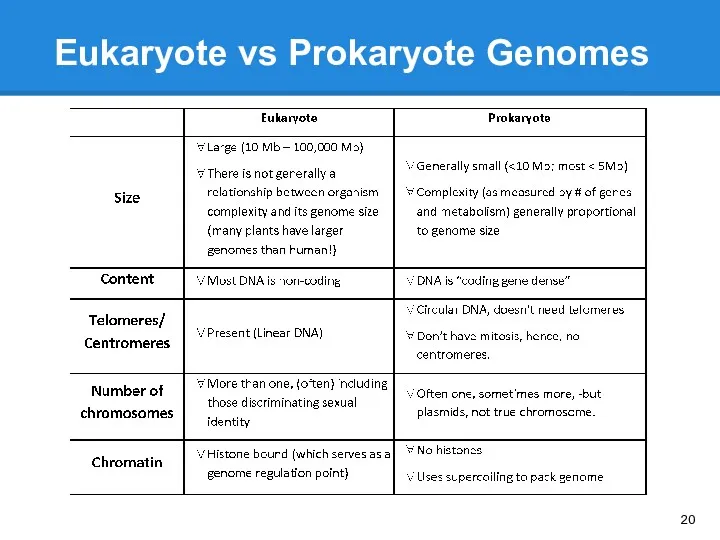
- 20. Eukaryote vs Prokaryote Genomes
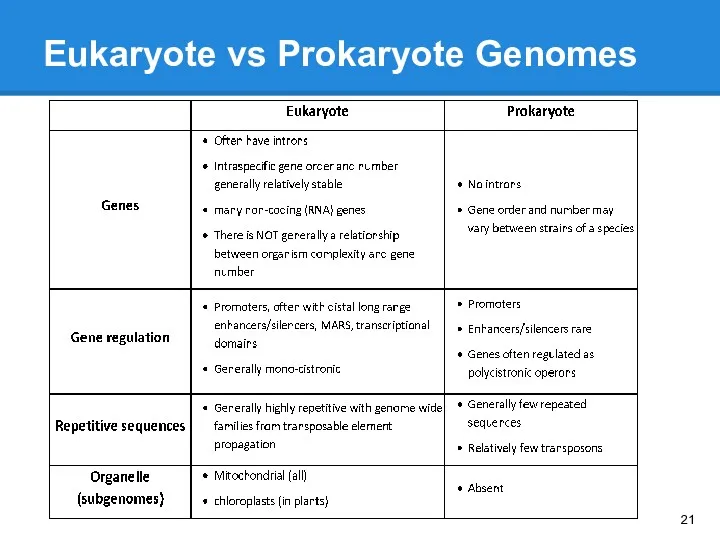
- 21. Eukaryote vs Prokaryote Genomes
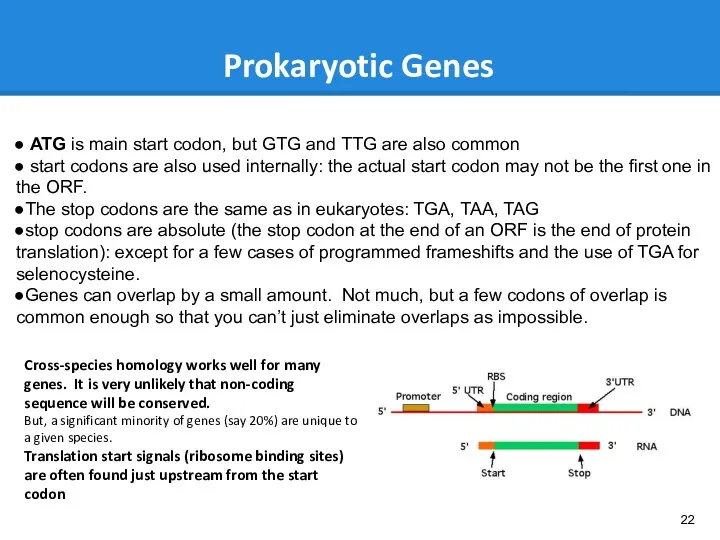
- 22. Prokaryotic Genes ATG is main start codon, but GTG and TTG are also common start codons
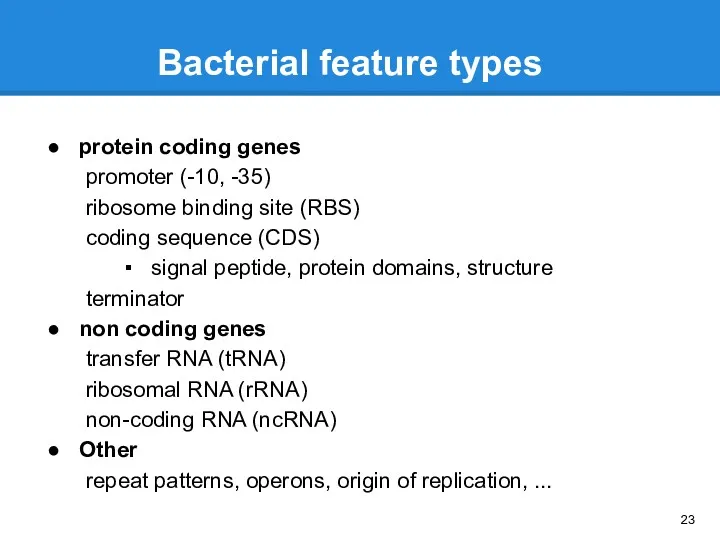
- 23. Bacterial feature types protein coding genes promoter (-10, -35) ribosome binding site (RBS) coding sequence (CDS)
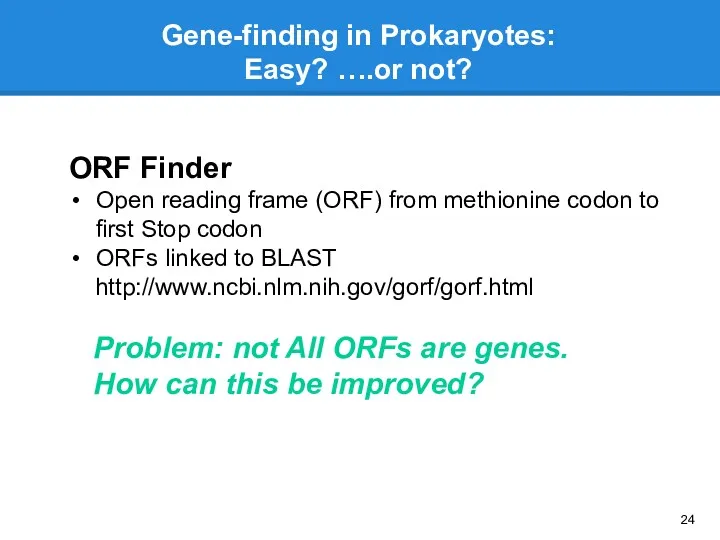
- 24. Gene-finding in Prokaryotes: Easy? ….or not? ORF Finder Open reading frame (ORF) from methionine codon to
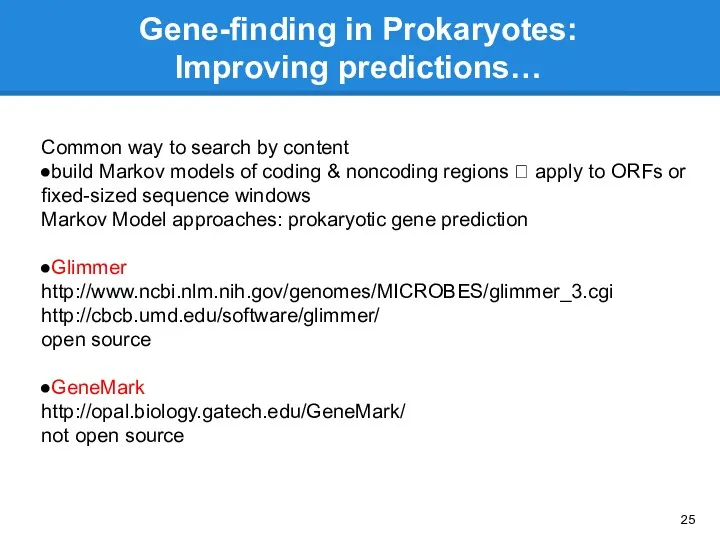
- 25. Gene-finding in Prokaryotes: Improving predictions… Common way to search by content build Markov models of coding
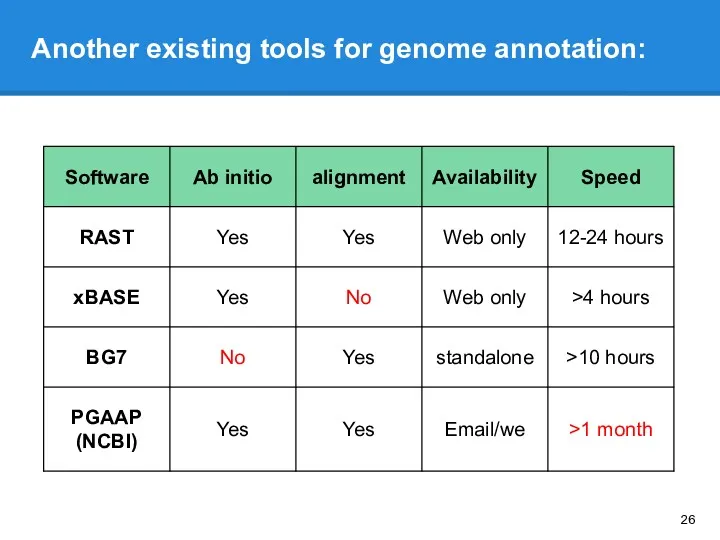
- 26. Another existing tools for genome annotation:
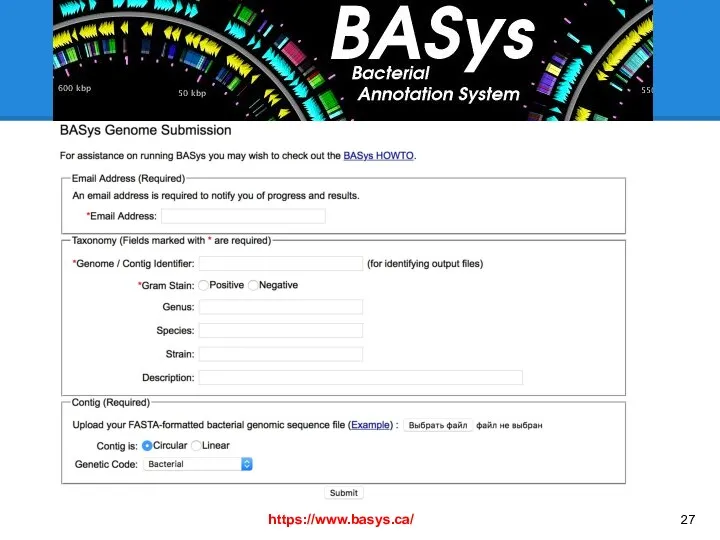
- 27. https://www.basys.ca/
- 28. designed for Bacteria, Archaea and Viruses. It can't handle multi-exon gene models your own custom "trusted"
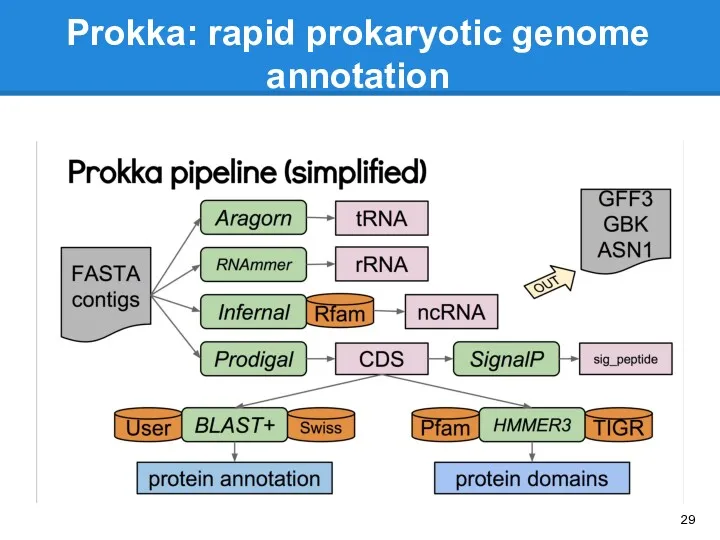
- 29. Prokka: rapid prokaryotic genome annotation
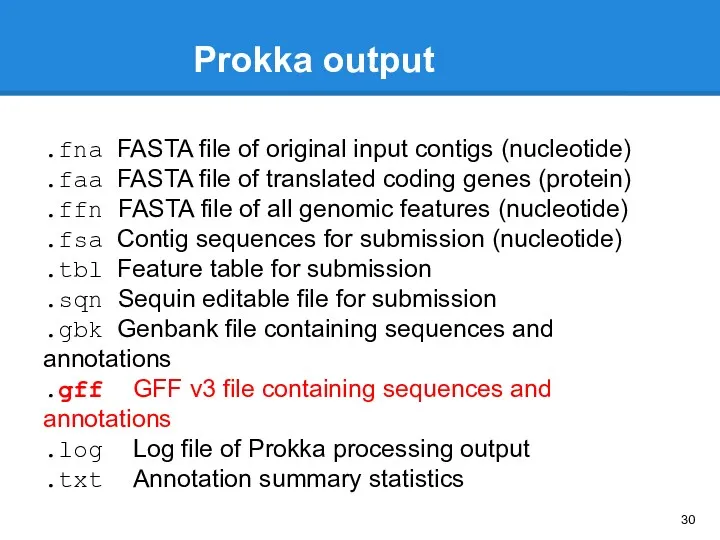
- 30. Prokka output .fna FASTA file of original input contigs (nucleotide) .faa FASTA file of translated coding
- 31. Prokka prokka --help prokka --docs Show full manual/documentation prokka --setupdb prokka --listdb List all configured databases
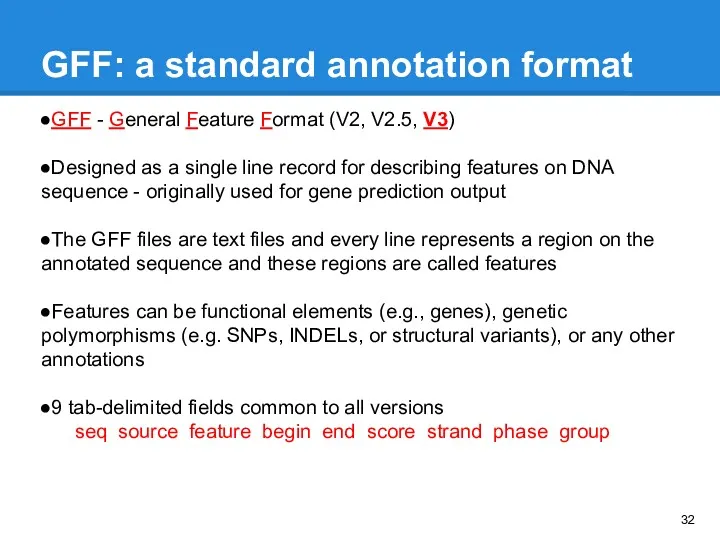
- 32. GFF - General Feature Format (V2, V2.5, V3) Designed as a single line record for describing
- 33. GFF-version 3 GROUP tag different for ALL versions GFF2: group is a unique description, usually the
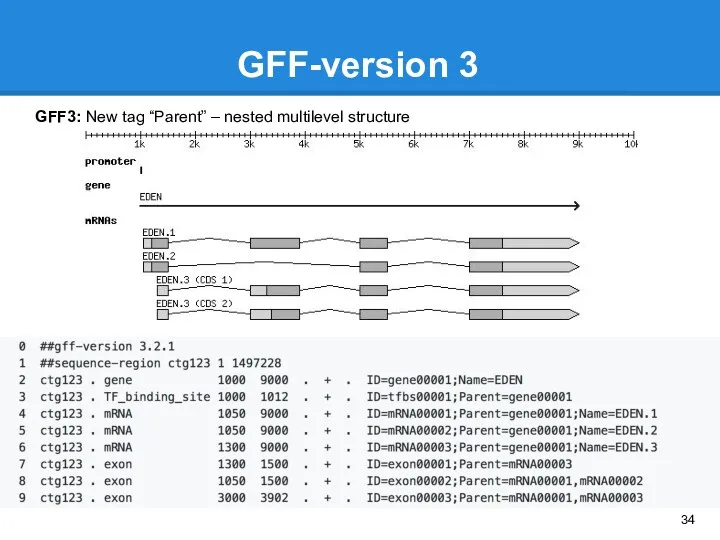
- 34. GFF-version 3 GFF3: New tag “Parent” – nested multilevel structure ctg123 . gene 1000 9000 .
- 35. GFF-version 3 GFF3: FASTA seqs can be embedded
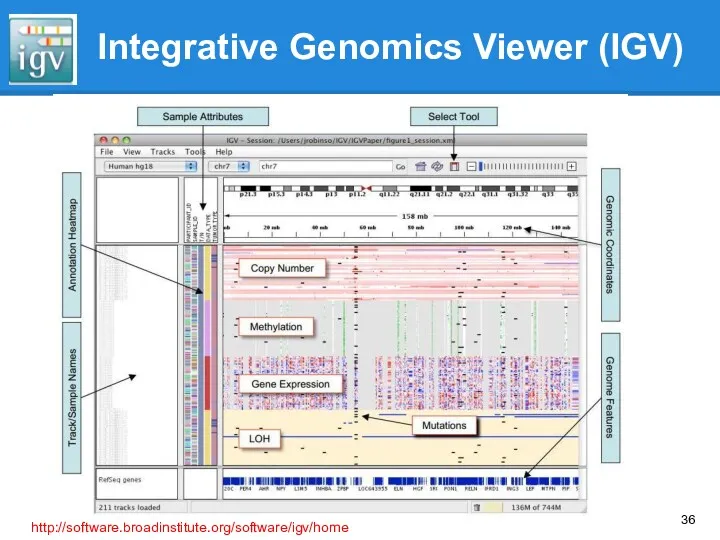
- 36. Integrative Genomics Viewer (IGV) http://software.broadinstitute.org/software/igv/home
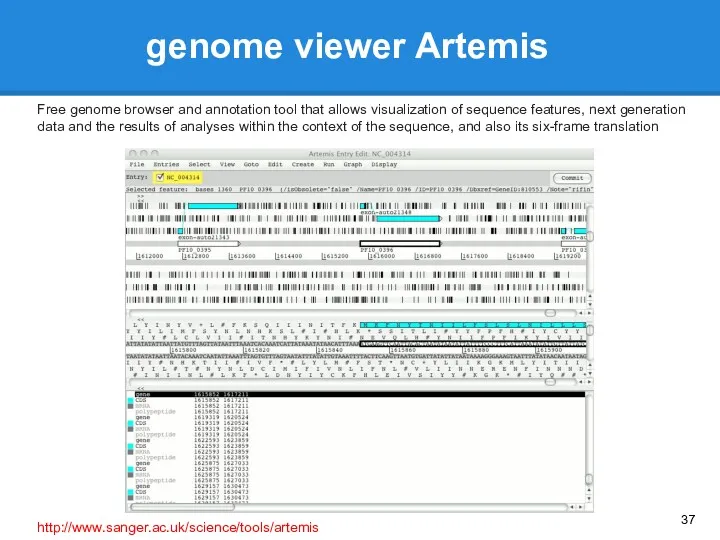
- 37. genome viewer Artemis Free genome browser and annotation tool that allows visualization of sequence features, next
- 39. Скачать презентацию









 Физиологический процесс дыхание
Физиологический процесс дыхание Общая характеристика надкласса Рыбы. Хрящевые рыбы
Общая характеристика надкласса Рыбы. Хрящевые рыбы Тыныс алудың организм үшін маңызы. Тыныс алу кезеңдері. Тыныс алудың реттелуі
Тыныс алудың организм үшін маңызы. Тыныс алу кезеңдері. Тыныс алудың реттелуі Ядовитые растения
Ядовитые растения Введение в клеточную биологию. Свойства живого. Клеточная теория
Введение в клеточную биологию. Свойства живого. Клеточная теория Надцарство актиномицеты
Надцарство актиномицеты Характеристика іонізуючих випромінювань і взаємодія їх із речовиною
Характеристика іонізуючих випромінювань і взаємодія їх із речовиною Зуби ссавців
Зуби ссавців Урок биологии в 9 классе по теме Антропогенез
Урок биологии в 9 классе по теме Антропогенез Селекция растений
Селекция растений Антропогенез. Расы. Расизм. Часть 5
Антропогенез. Расы. Расизм. Часть 5 Клиническая анатомия органов малого таза у женщин
Клиническая анатомия органов малого таза у женщин Исчезающие виды птиц в Самарской области
Исчезающие виды птиц в Самарской области Слюнные железы
Слюнные железы Структурно-функциональная организация клетки
Структурно-функциональная организация клетки Класс Пресмыкающиеся, или Рептилии. Отряд Чешуйчатые
Класс Пресмыкающиеся, или Рептилии. Отряд Чешуйчатые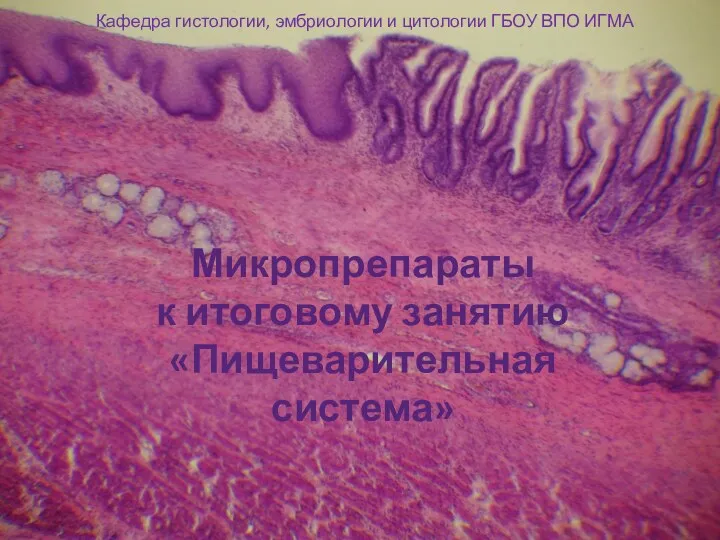 Пищеварительная система. Микропрепараты по гистологии
Пищеварительная система. Микропрепараты по гистологии Жасуша түралы түсінік
Жасуша түралы түсінік Гюннинен 211 мутагены
Гюннинен 211 мутагены Хвощи. Внешний вид хвоща
Хвощи. Внешний вид хвоща Растительный и животный мир Антарктиды
Растительный и животный мир Антарктиды Өсімдіктер
Өсімдіктер Вегетативное размножение.
Вегетативное размножение. Лекарственные растения
Лекарственные растения Науки об организме человека
Науки об организме человека Причини вимирання динозаврів
Причини вимирання динозаврів Птицы леса
Птицы леса Презентация Биография И. Н. Сеченова, 8 кл
Презентация Биография И. Н. Сеченова, 8 кл