Содержание
- 2. Approaches to problems in cell biology Biochemistry-You can define a enzyme reaction (protein) and then try
- 3. Resolution of instruments in cellular biology Resolution describes the minimal distance of two points that can
- 4. Resolution of instruments in cell biology (2 objects) Visible light is 400-700nm Dry lens(0.5NA), green(530nm light)=0.65µm=650nm
- 5. Sizes of objects Eukaryotic cell- 20µm Procaryotic cell-1-2µm nucleus of cell-3-5µm mitochondria/chloroplast- 1-2µm ribosome- 20-30nm protein-
- 6. Basic info expected from flow cytometry experiment (2 cellular populations): . Whether a cell of interest
- 7. Analysis of Cellular subpopulations by different methods (How many parameters to measure?) Conventional flow cytometry -4---12---18
- 8. Speed and Statistics (How fast? How precise?) Microscopy (20x-100x objective) – 20-100 cells/per slide or well
- 9. Zeno’s paradox
- 10. STATISTICS: How many cells we really need to count?
- 11. It depends from heterogeneity of cell population, % of antigen expression etc etc
- 12. File size for Imagestream imaging flow cytometer –up to 100,000 events (cell images) allows to work
- 13. Basic Flow Cytometer How does it work? Fluidics (stream) Optics/excitation sources Electronics Fluidics Hydrodynamic focusing of
- 14. Sample Injection port: sheath flow laser beams Hydrodynamic focusing Sample core stream Sample cells at interrogation
- 15. Optics:Light Sources Light Amplification by the Stimulated Emission of Radiation s Can provide a single wavelength
- 16. Solid state lasers-small, reliable, easy to integrate in existing technology and are rapidly decreasing in the
- 17. Multiple lasers in modern flow cytometer LSR2 7 lasers LSRFortessa 5 lasers Influx… 6 lasers Stratedigm
- 18. FACS Aria sorter
- 19. FACSCalibur flow cytometer
- 20. Optics: Forward Scatter Channel/Side Scatter Channel FSC influenced by particle size and shape; Allows the computer
- 21. Light Scattering properties of cells Right Angle Light Detector α Cell Complexity Forward Light Detector α
- 22. Neutrophils Monocytes Lymphocytes Forward Light Scatter Analyze (gate on) cells of interest Lysed Whole Blood Side
- 24. Scatter (Size parameter)-by conventional flow cytometry and IFC Barteneva et al, BBA Reviews on Cancer 2013,
- 25. Principle of fluorescence Principle of Fluorescence 1. Energy is absorbed by the atom which becomes excited.
- 26. FLUORESCENT methods in the research laboratory State-of-the art Fluorescent Microscopy and Confocal Microscopy High dimensional Flow
- 27. Advantages of fluorescent methods Highly sensitive method (high resolution) Highly sophisticated fluorescent probes (multi-) Fluorescent dyes
- 28. FLUORESCENT dyes are typically composed of ring structures
- 29. Absorption and Emission Spectra of some traditional fluorophores
- 30. Fluorescence Stoke’s shift Fluorescence emission peak wavelength is red-shifted with respect to absorption peak wavelength This
- 31. USE OF FLUORESCENT DYES Labeling of proteins - antibodies, streptavidin Labeling of nucleic acids – DNA,
- 32. FITC (Fluorescein isothiocyanate) Fluorescein isothiocyanate is a yellow-green colored low molecular weight dye which couples to
- 33. R-PE - R symbolises its red-algal origin – it is a bright orange-red colored protein, with
- 34. APC(allophycocyanin) Rod-and-core structure of cyanobacterial phycobilisome. Left-hand diagram shows stacks of hexameric phycocyanin complexes comprising the
- 35. ALEXA family:brighter, more photostable, less environmental sensitive
- 36. Quantum Dot-conjugated antibody
- 37. Quantum Dots advantages Extremly photostable Narrow emission spectrum, hence small spectral overlap Broad absorption spectrum (
- 38. QDots Brightness Brightness Index=Extinction Coefficient x Quantum Yield/1000
- 39. How do we get fluorescent probes into cells Kill the cell and make the membrane permeable
- 40. How to load cells (microscopy) .
- 41. Immunofluorescent staining of proteins in fixed/dead cells You can purify almost any protein from the cell
- 42. Protein from fluorescent jellyfish The protein is fluorescent Now cloned, sequenced and X-ray structure known If
- 43. Discovery of fluorescent proteins
- 44. Evrogen proteins (Lukianov Lab)
- 45. Conventional flow cytometry (Example: scattering+5 colors)
- 46. 9 colors: Murine Hematopoietic Stem Cells Sort from Transplant Objective: To serially transplant subpopulations of hematopoietic
- 47. Imaging flow cytometers provide alternative for cellular analysis and characterization
- 48. Imagestream 100 imaging flow cytometer
- 49. TDI CCD Excite fluorescence over the entire height of the detector Light is detected in the
- 50. Imagestream X Mark II x60 objective; higher acquisition speed; 10 fluorescent channels; +561 nm laser
- 51. Imagestream X Mark II Amnis Inc
- 52. Imagestream (s) optical configurations and fluorescent channels Adapted from A.Filby, 2015
- 53. Cellular analysis by conventional Flow Cytometry Traditional markers to define cell populations (human, rat, mouse) Relies
- 54. Standard approach to verify FACS-defined cellular subset:cell sorting+microscopy From Becher et al, Nature Immunology, 2014
- 55. Limitations of FACS sorting/microscopy approach Purity of sorted subpopulation (never 100%)-can be 85% or less for
- 56. Aspect Ratio Intensity is the minor axis intensity divided by the major axis intensity. Identifying single
- 57. Shape parameters in defining erythroid sickle anemia cells (Samsel, McCoy Jr, 2016)
- 58. Size/Shape distribution analysis (Aphanizomenon sp. Cells, our data)
- 59. Fluorescence-based analysis by Imagestream DNA/RNA dyes (PI, Sytox Blue, SYTOX Green etc) Lipid dyes (DiO, DiA,
- 60. Description: Intensity is the sum of all the pixel values in the mask, background subtracted. Intensity:
- 61. Quantification of Toxoplasma gondii Muskavitch et al, 2008
- 62. Number of ingested by neutrophils S. aureus bacteria (Ploppa et al, 2011)
- 63. Counting of Leishmania donovani (% infected cells and #parasites/cell) (Torrezas et al, 2015)
- 64. Internalization of CSFE-stained N.gonorrhoeae bacteria (Smirnov et al, 2015)
- 65. Human PBMC -morphology (from B.Hall)
- 66. AMNIS CORPORATION-Compensation Single color control samples used to calculate a 6x6 matrix. Post-acquisition compensation is applied
- 67. Spectral compensation is assymetric
- 68. From 3-4 colors for images (microscopy) to 8-colors immunophenotyping (external staining) with Imagestream X Mark II
- 69. Untranslocated Translocated NFkB Translocation Using The Similarity Algorithm (Amnis)
- 70. Untranslocated Translocated NFkB Translocation Using The Similarity Algorithm (Amnis)
- 71. Bystander MFs have impaired NFkappaBeta translocation to the nucleus (Torrez et al, 2015)
- 72. Co-localisation
- 73. Case 1: Co-localisation M.tuberculosis with Rab5 and Rab7 (From Haridas et al, 2016)
- 74. Co-localisation of S.aureus/dihydroethidium (oxidative burst in human whole blood) (Ploppa et al, 2011)
- 75. Nuclear fragmentation/caspase activity
- 77. Скачать презентацию










































































 Клиническая биохимия азотистого обмена. (Лекция 7)
Клиническая биохимия азотистого обмена. (Лекция 7) Органы растений. Цветок
Органы растений. Цветок Членистоногие
Членистоногие 5.02- 7кл внутреннее строение листа_
5.02- 7кл внутреннее строение листа_ Мутационная и комбинативная изменчивость
Мутационная и комбинативная изменчивость Мейоз. Типы мейоза
Мейоз. Типы мейоза Общая характеристика отдела моховидные
Общая характеристика отдела моховидные Structura celulei bacteriene
Structura celulei bacteriene Задание № 27 (ОГЭ)
Задание № 27 (ОГЭ) Дикорастущие растения и цветы
Дикорастущие растения и цветы Биологические ритмы
Биологические ритмы Исследовательская деятельность на уроках биологии и во внеурочное время как средство развития творческих способностей
Исследовательская деятельность на уроках биологии и во внеурочное время как средство развития творческих способностей Витамины
Витамины Protein synthesis
Protein synthesis Химический состав растений
Химический состав растений Нуклеиновые кислоты и их роль в жизнедеятельности клетки
Нуклеиновые кислоты и их роль в жизнедеятельности клетки 5 весняних квіток
5 весняних квіток Строение клетки
Строение клетки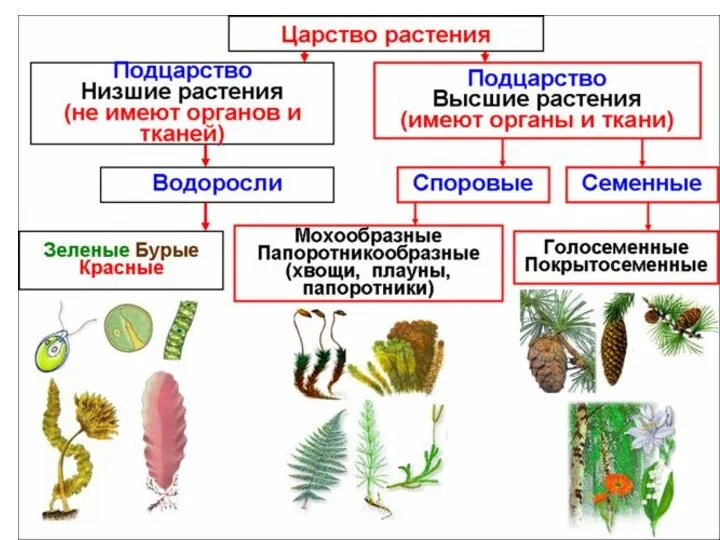 Царство растения
Царство растения Тип Моллюски
Тип Моллюски Опасные животные России
Опасные животные России Вирусы – неклеточные формы жизни
Вирусы – неклеточные формы жизни Содержание углекислого газа в школьных помещениях
Содержание углекислого газа в школьных помещениях Грибы, травы, цветы
Грибы, травы, цветы Нервная система человека
Нервная система человека УРОК Образ мира
УРОК Образ мира Зоология позвоночных. Строение основных типов беспозвоночных
Зоология позвоночных. Строение основных типов беспозвоночных Бізон європейський, або зубр
Бізон європейський, або зубр