Содержание
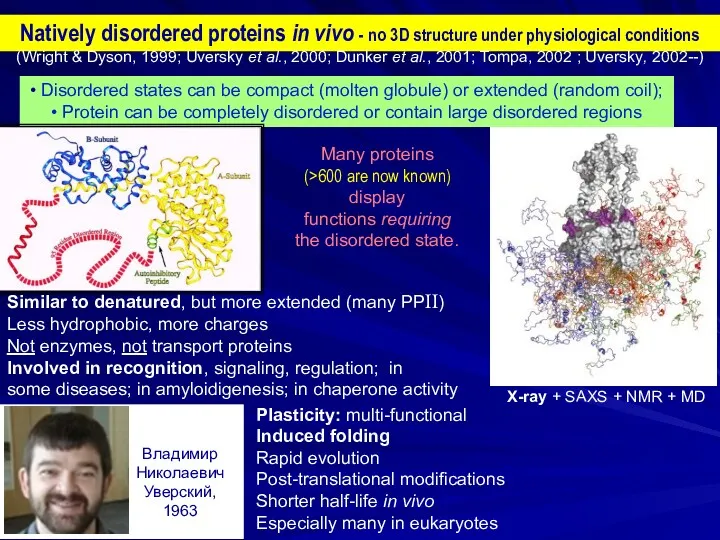
- 2. Natively disordered proteins in vivo - no 3D structure under physiological conditions • Disordered states can
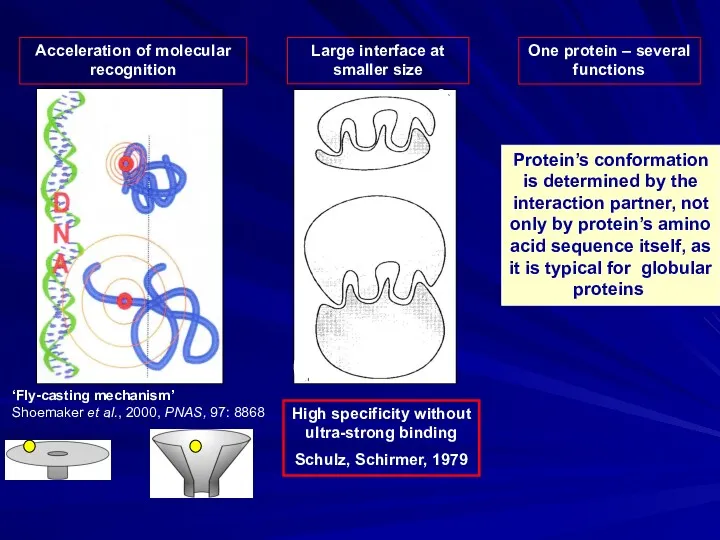
- 3. Acceleration of molecular recognition One protein – several functions Protein’s conformation is determined by the interaction
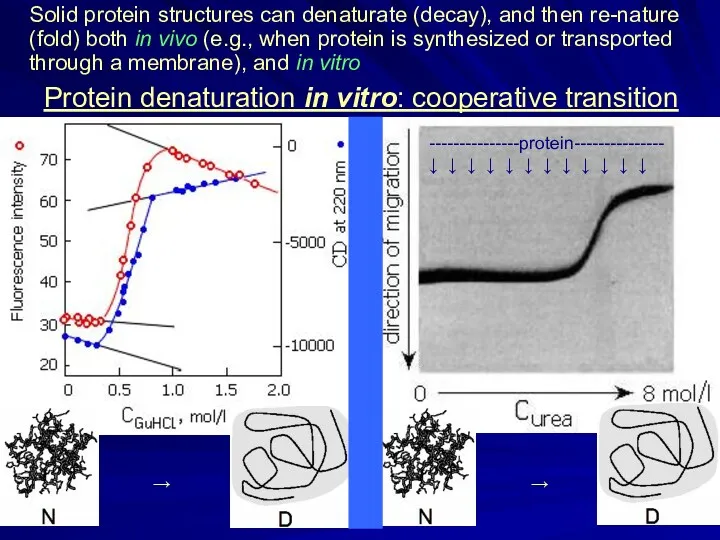
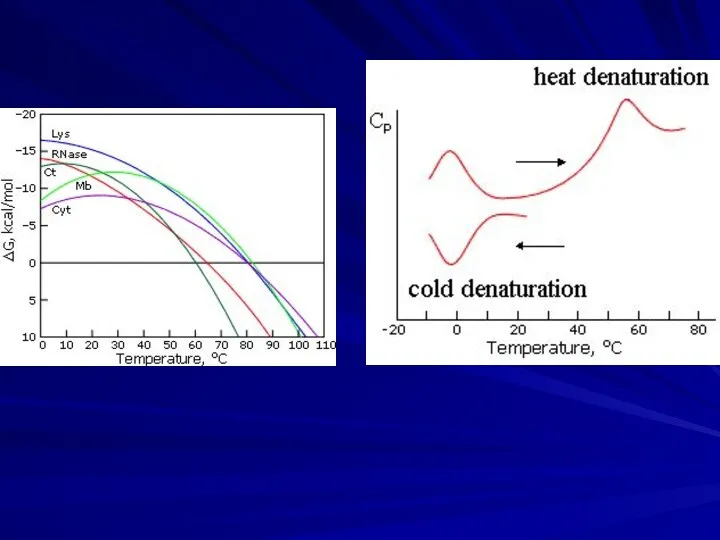
- 4. Protein denaturation in vitro: cooperative transition Solid protein structures can denaturate (decay), and then re-nature (fold)
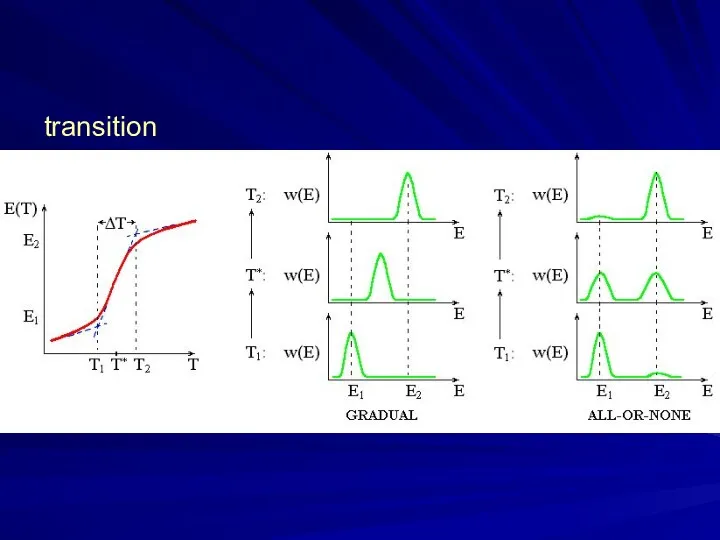
- 5. transition
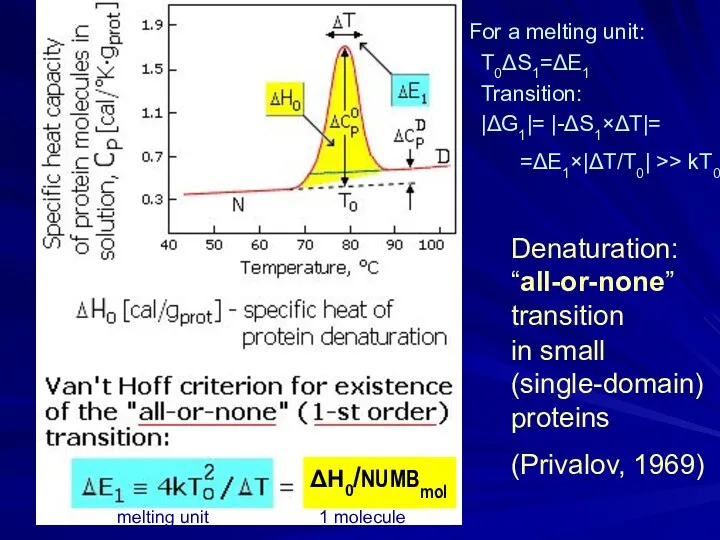
- 6. Denaturation: “all-or-none” transition in small (single-domain) proteins (Privalov, 1969) For a melting unit: T0ΔS1=ΔE1 Transition: |ΔG1|=
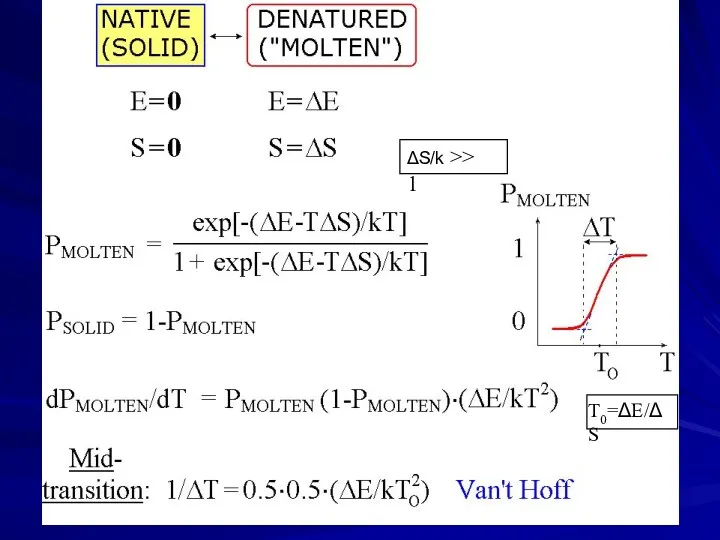
- 7. ΔS/k >> 1 T0=ΔE/ΔS
- 8. Jacobus Henricus van 't Hoff, Jr. (1852 –1911) The first Nobel prize in Chemistry, 1901 ПРИВАЛОВ
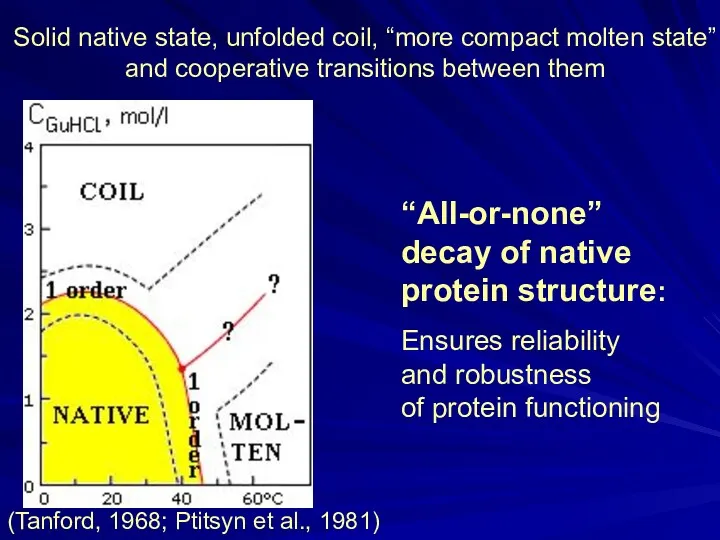
- 10. “All-or-none” decay of native protein structure: Ensures reliability and robustness of protein functioning Solid native state,
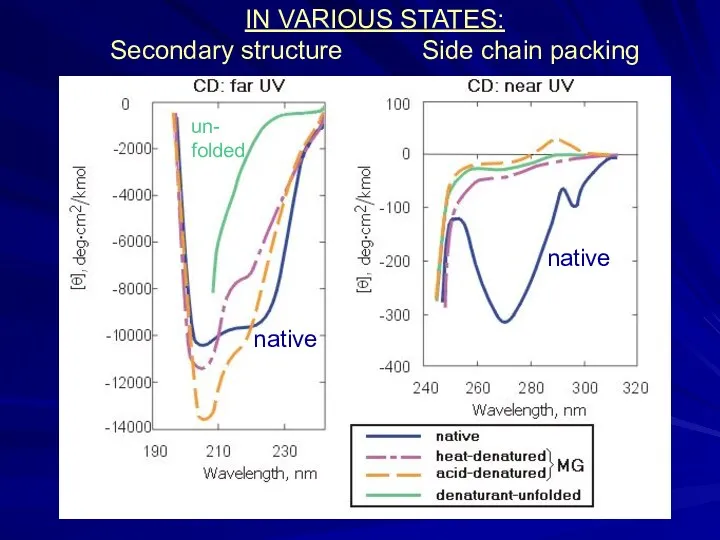
- 11. IN VARIOUS STATES: Secondary structure Side chain packing native un- folded native
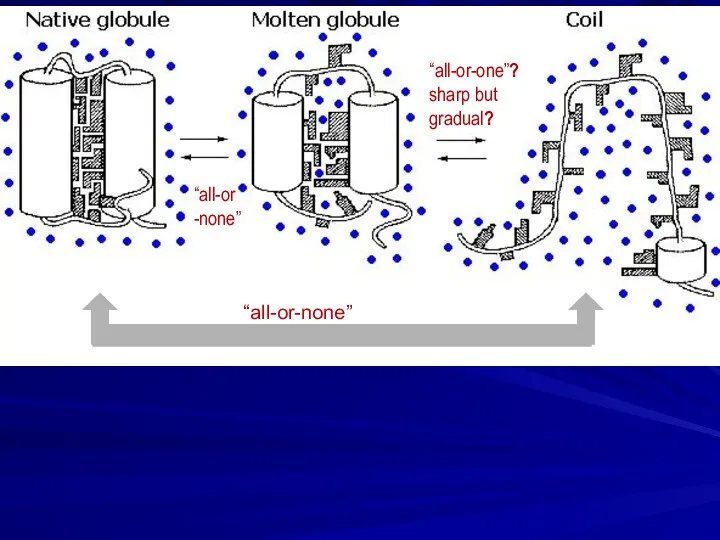
- 12. “all-or-none” “all-or -none” “all-or-one”? sharp but gradual?
- 13. Евгений Исаакович Шахнович, 1957 Дмитрий Александрович Долгих, 1954 Геннадий Васильевич Семисотнов, 1947 Олег Борисович Птицын (1929-99)
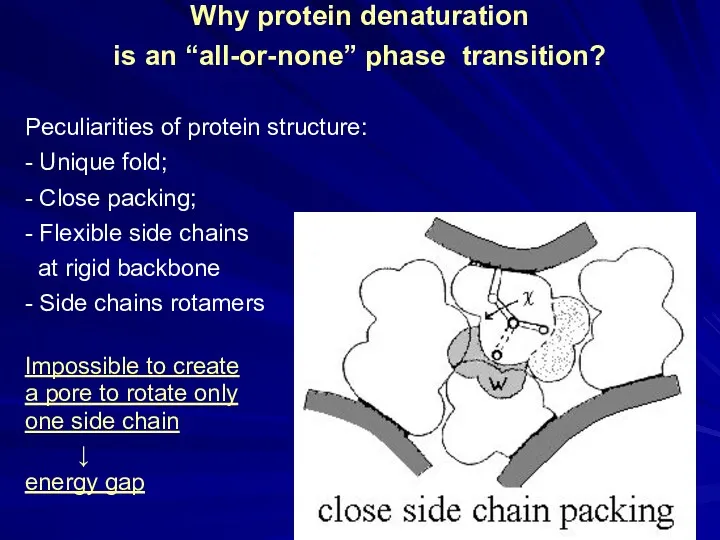
- 14. Why protein denaturation is an “all-or-none” phase transition? Peculiarities of protein structure: - Unique fold; -
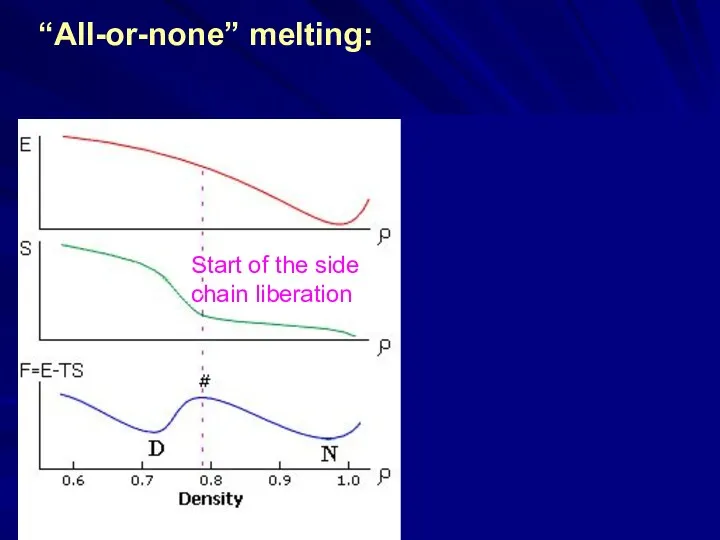
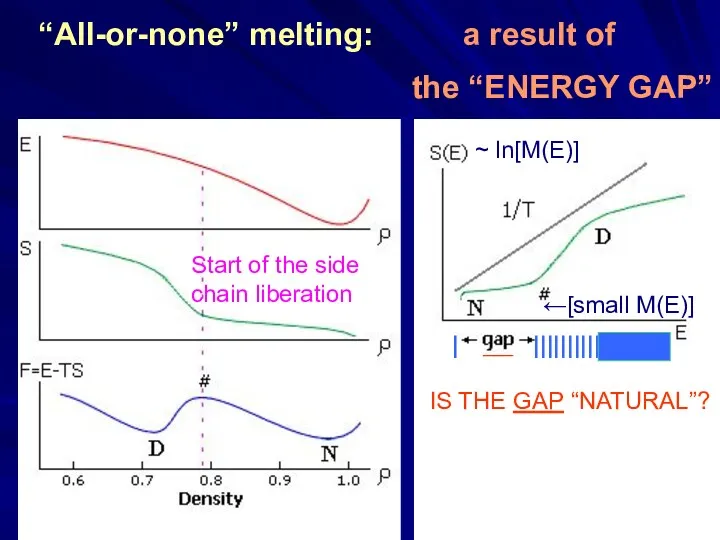
- 15. Start of the side chain liberation “All-or-none” melting:
- 16. “All-or-none” melting: a result of the “ENERGY GAP” Start of the side chain liberation ~ ln[M(E)]
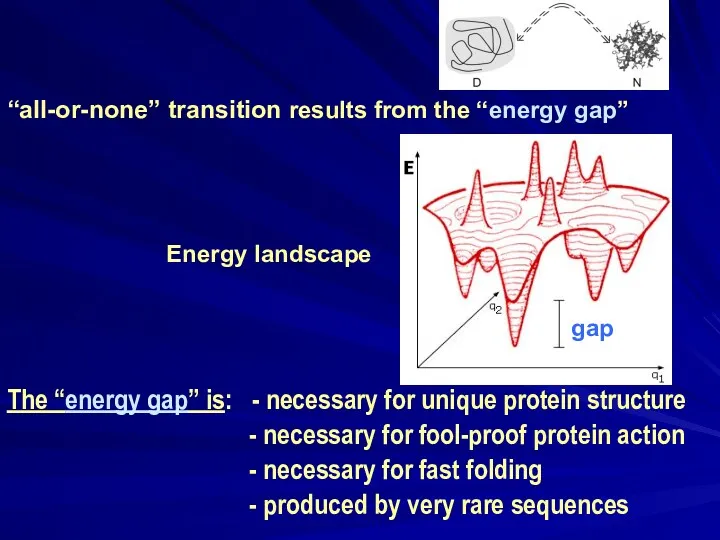
- 17. “all-or-none” transition results from the “energy gap” Energy landscape The “energy gap” is: - necessary for
- 18. GAP WIDTH: MAIN PROBLEM OF EXPERIMENTAL PROTEIN PHYSICS PHYSICAL ESTIMATE: =??? BIOLOGICAL ESTIMATE: 1 0F ~1010
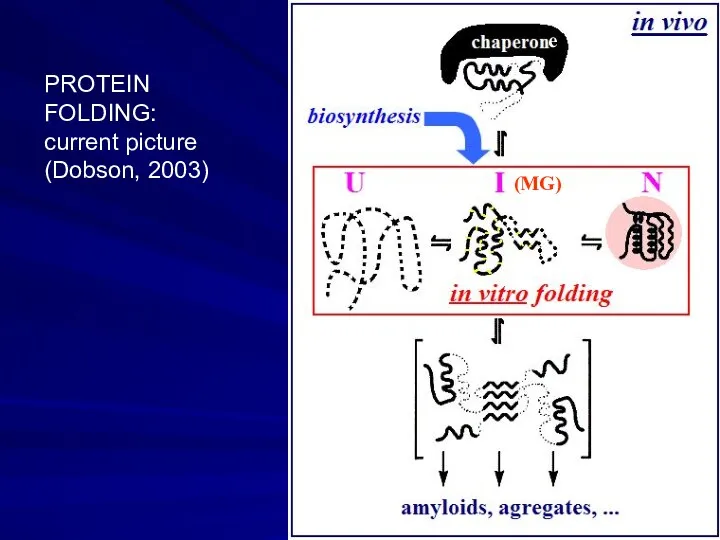
- 19. e PROTEIN FOLDING: current picture (Dobson, 2003) (MG)
- 21. Скачать презентацию



 Значение влажности в жизни человека
Значение влажности в жизни человека 26e1e5eed06e9616d6b0b1a82d75cb58
26e1e5eed06e9616d6b0b1a82d75cb58 Area, size and mass
Area, size and mass Постоянные магниты (8 класс)
Постоянные магниты (8 класс) Инфракрасное, ультрафиолетовое и рентгеновское излучения. Их свойства и применение
Инфракрасное, ультрафиолетовое и рентгеновское излучения. Их свойства и применение Физика и техника
Физика и техника Проект Инженерный класс в московской школе. Практические ситуационные задачи и теоретические задачи
Проект Инженерный класс в московской школе. Практические ситуационные задачи и теоретические задачи Законы постоянного тока
Законы постоянного тока Основні уявлення та рівняння
Основні уявлення та рівняння Баяндама Альберт Эйнштейн
Баяндама Альберт Эйнштейн Векторный анализ и синтез сигналов. Программа “Вектор”
Векторный анализ и синтез сигналов. Программа “Вектор” Подготовка к ОГЭ по физике на уроке. Решение задач по физике различного типа и уровня сложности
Подготовка к ОГЭ по физике на уроке. Решение задач по физике различного типа и уровня сложности Tucson (NX 4). Зависание клапана CVVT
Tucson (NX 4). Зависание клапана CVVT Теплопередача. Задачи
Теплопередача. Задачи Физика атома и атомных явлений
Физика атома и атомных явлений Сила. Явление тяготения. Сила тяжести
Сила. Явление тяготения. Сила тяжести История развития нанотехнологий
История развития нанотехнологий Алгоритмы решения физических задач
Алгоритмы решения физических задач Unusual modes pf transport
Unusual modes pf transport Виды теплопередачи ( презентация к уроку)- 8 класс
Виды теплопередачи ( презентация к уроку)- 8 класс Получение и передача переменного электрического тока. Трансформатор
Получение и передача переменного электрического тока. Трансформатор Состав систем космического аппарата. Проведение литературного обзора
Состав систем космического аппарата. Проведение литературного обзора Основы машиноведения
Основы машиноведения Викторина по физике
Викторина по физике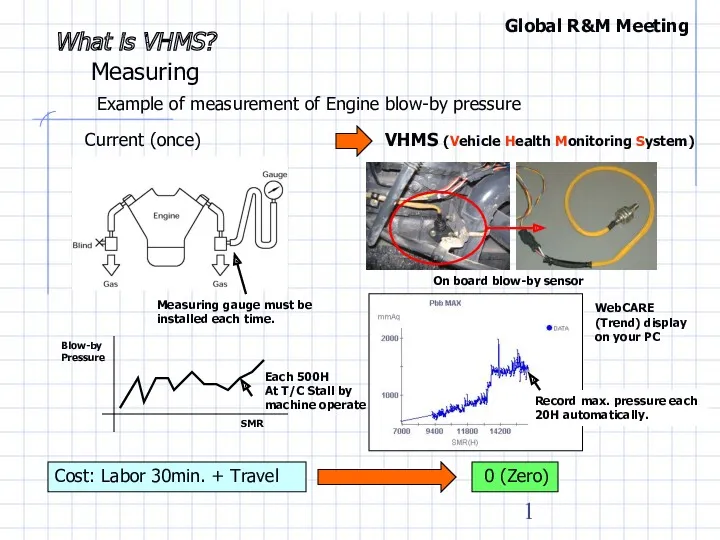 Summary of effective use 6
Summary of effective use 6 Основы квантовой физики. Лазеры
Основы квантовой физики. Лазеры Свойства воздуха
Свойства воздуха Система питания двигателя от впрыска топлива
Система питания двигателя от впрыска топлива