Содержание
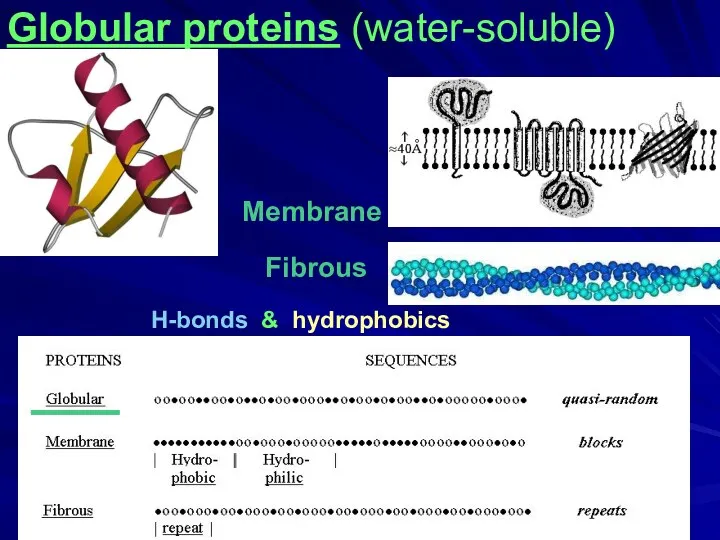
- 2. Fibrous H-bonds & hydrophobics Membrane ____ Globular proteins (water-soluble)
- 3. Hermann Emil Louis Fischer (1852 –1919) Nobel Prize 1902 Protein chain Protein sequence Frederick Sanger (1918
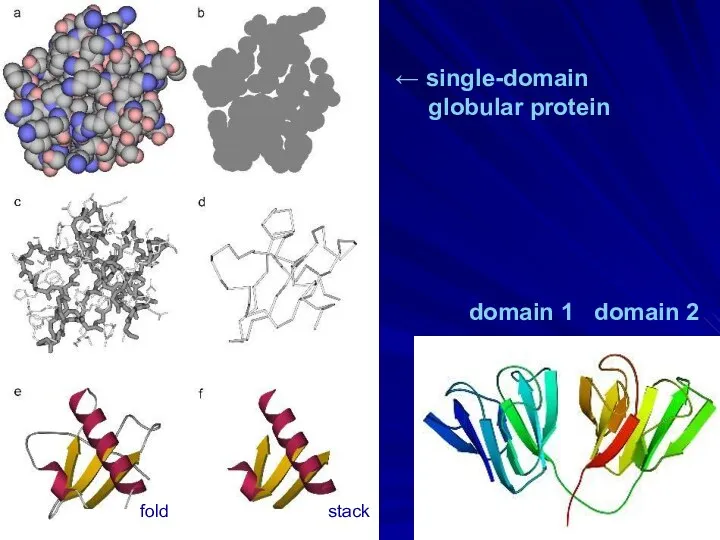
- 4. domain 1 domain 2 ← single-domain globular protein fold stack
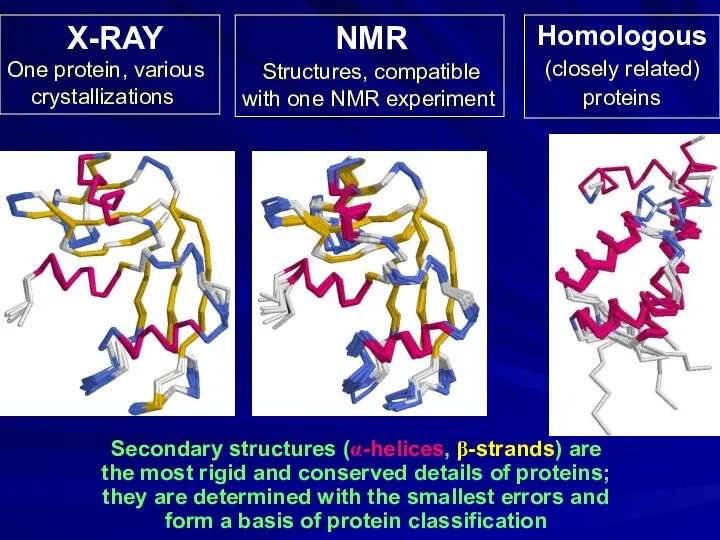
- 5. Secondary structures (α-helices, β-strands) are the most rigid and conserved details of proteins; they are determined
- 6. Max Ferdinand Perutz (1914 –2002) Nobel Prize 1962 X-ray 3D protein structure Kurt Wüthrich, 1938 Nobel
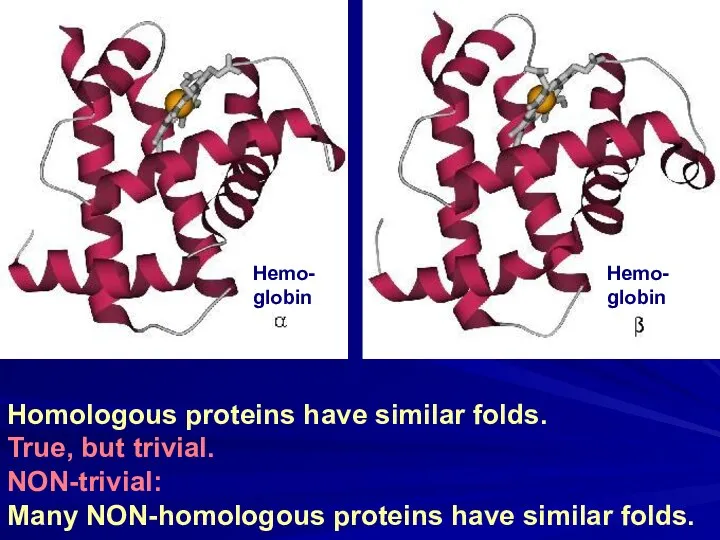
- 7. Homologous proteins have similar folds. True, but trivial. NON-trivial: Many NON-homologous proteins have similar folds. Hemo-
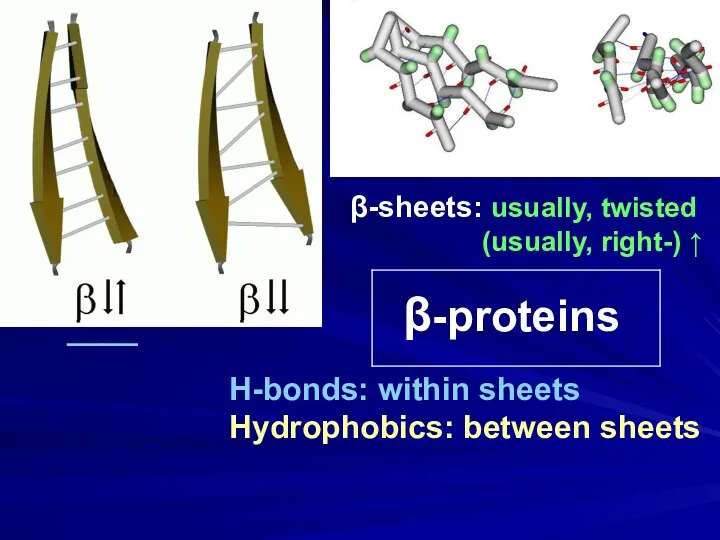
- 8. β-proteins β-sheets: usually, twisted (usually, right-) ↑ H-bonds: within sheets Hydrophobics: between sheets ____
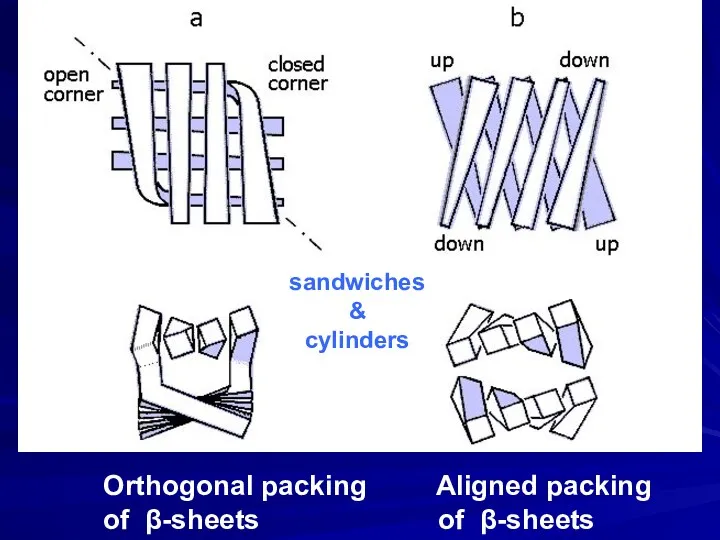
- 9. Orthogonal packing Aligned packing of β-sheets of β-sheets sandwiches & cylinders
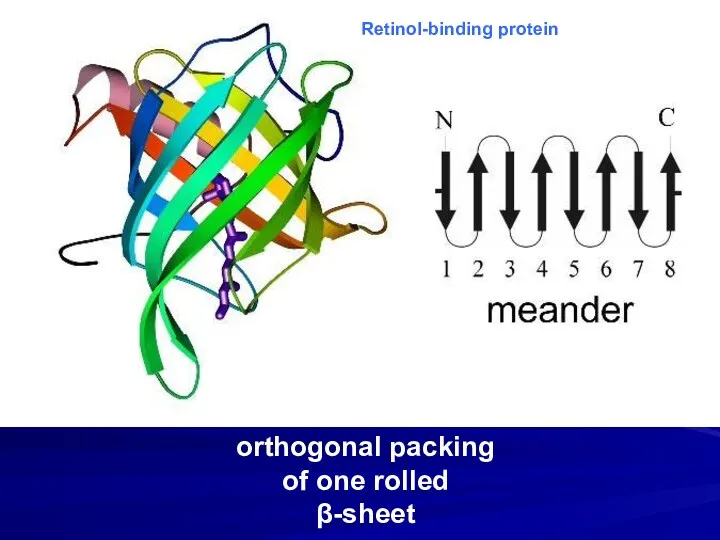
- 10. orthogonal packing of one rolled β-sheet Retinol-binding protein
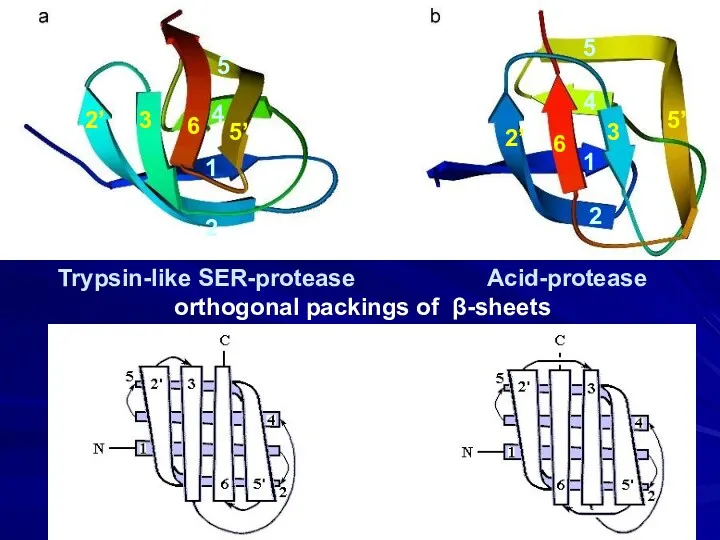
- 11. Trypsin-like SER-protease Acid-protease orthogonal packings of β-sheets 2 1 4 5 5’ 6 3 2’ 2
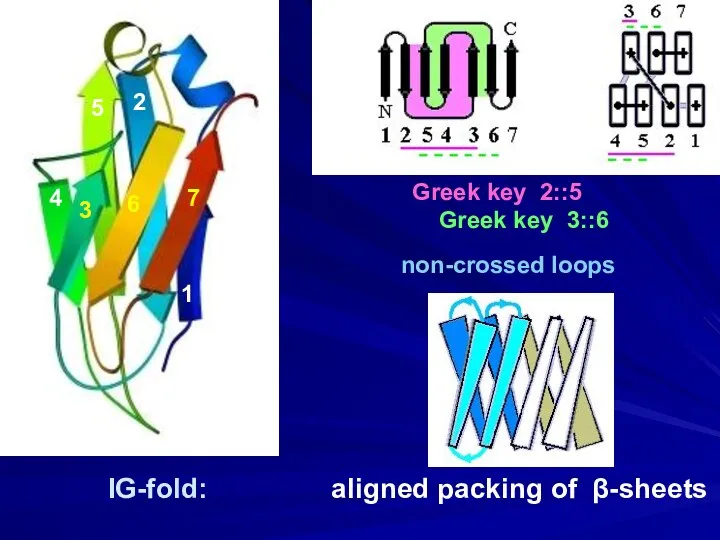
- 12. IG-fold: aligned packing of β-sheets Greek key 2::5 Greek key 3::6 1 2 3 4 5
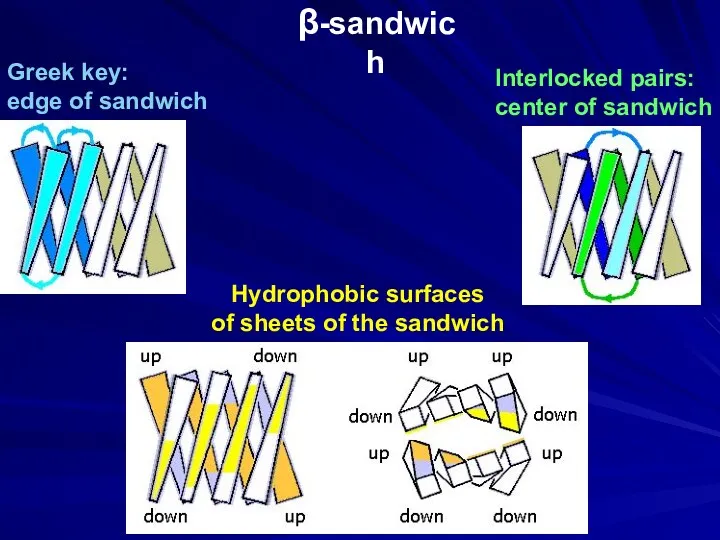
- 13. β-sandwich Interlocked pairs: center of sandwich Greek key: edge of sandwich Hydrophobic surfaces of sheets of
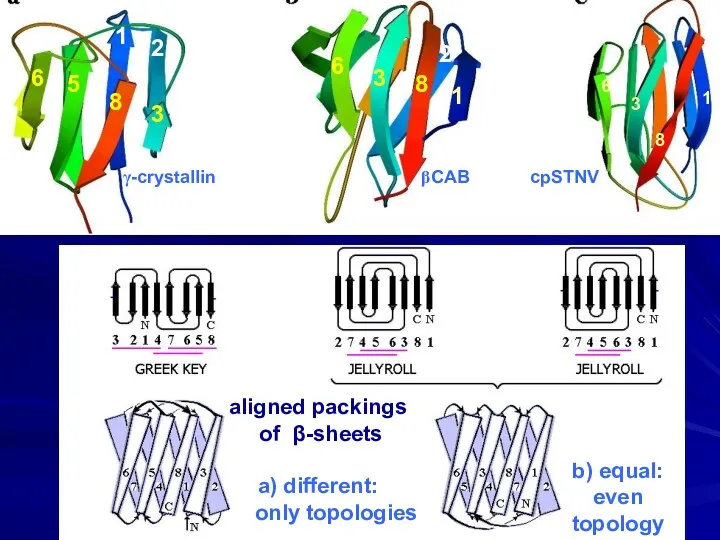
- 14. aligned packings of β-sheets a) different: only topologies b) equal: even topology 6 5 8 3
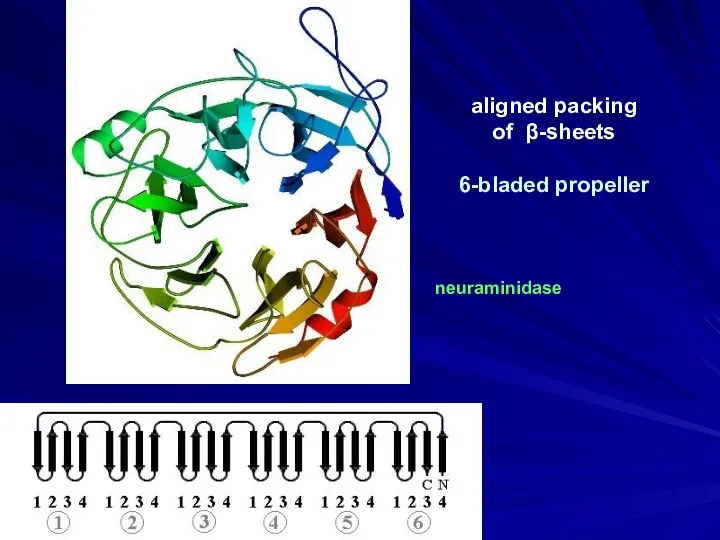
- 15. aligned packing of β-sheets 6-bladed propeller neuraminidase
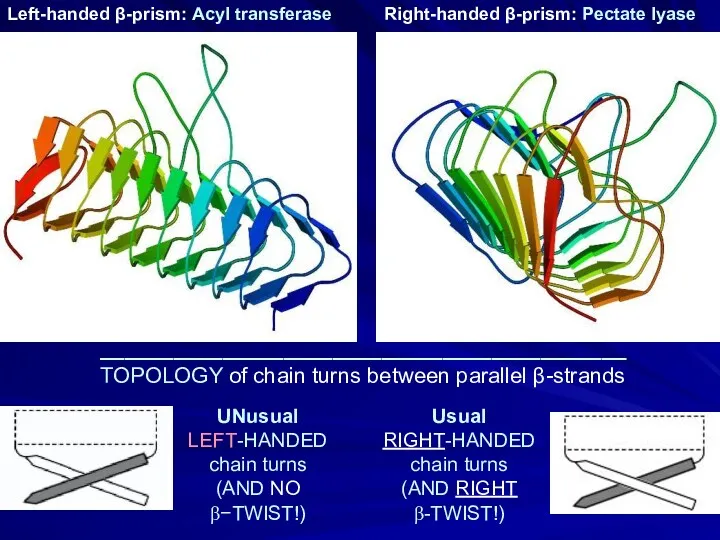
- 16. UNusual LEFT-HANDED chain turns (AND NO β−TWIST!) Left-handed β-prism: Acyl transferase Right-handed β-prism: Pectate lyase Usual
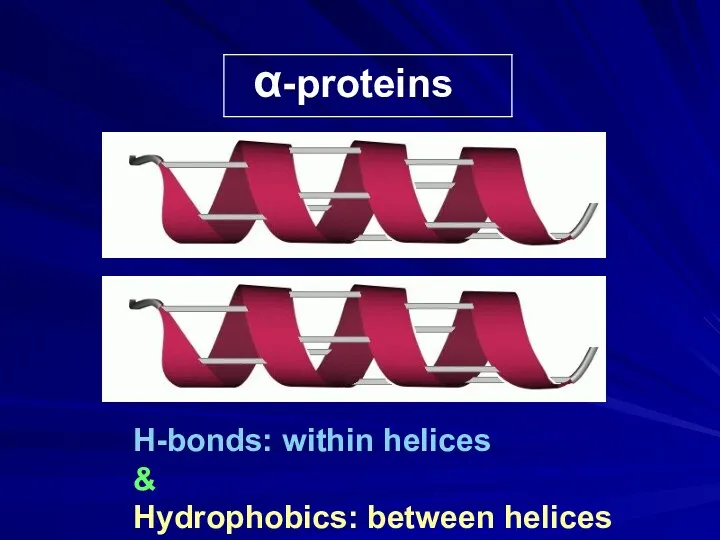
- 17. α-proteins H-bonds: within helices & Hydrophobics: between helices
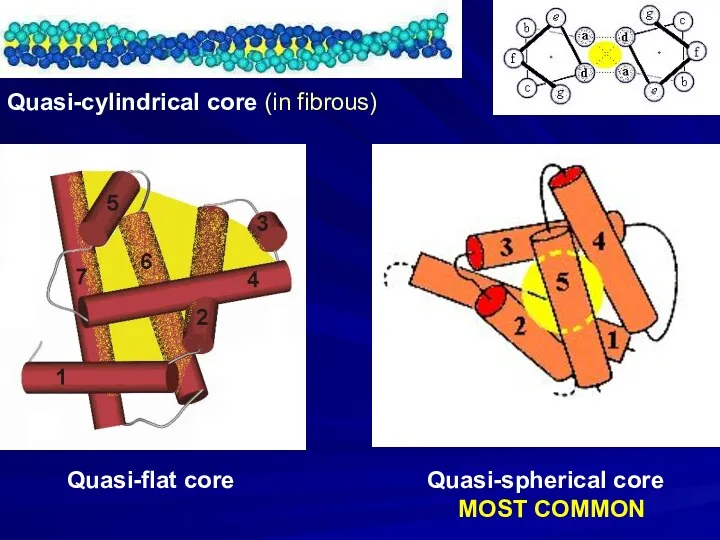
- 18. Quasi-cylindrical core (in fibrous) Quasi-flat core Quasi-spherical core MOST COMMON
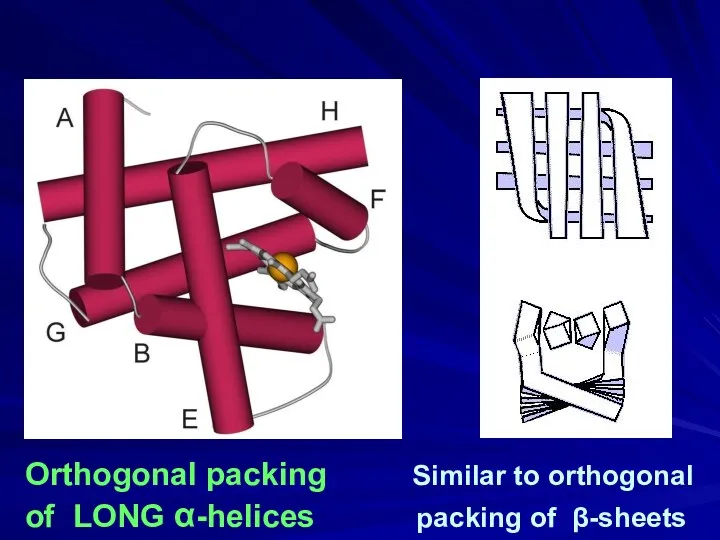
- 19. Orthogonal packing Similar to orthogonal of LONG α-helices packing of β-sheets
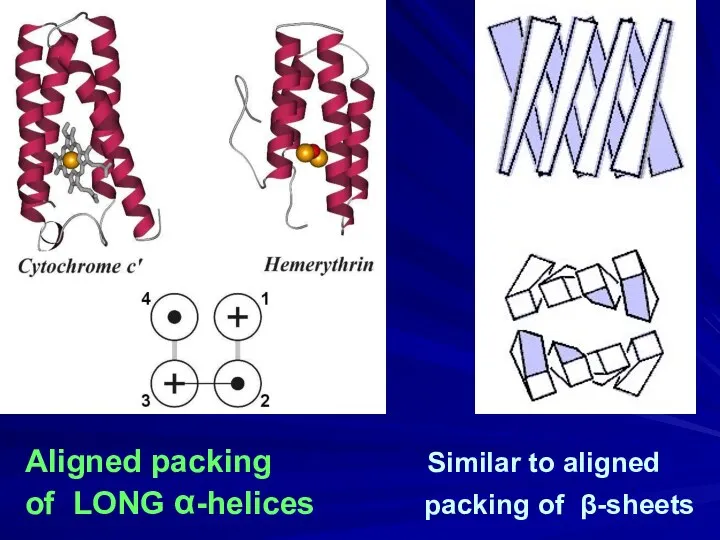
- 20. Aligned packing Similar to aligned of LONG α-helices packing of β-sheets
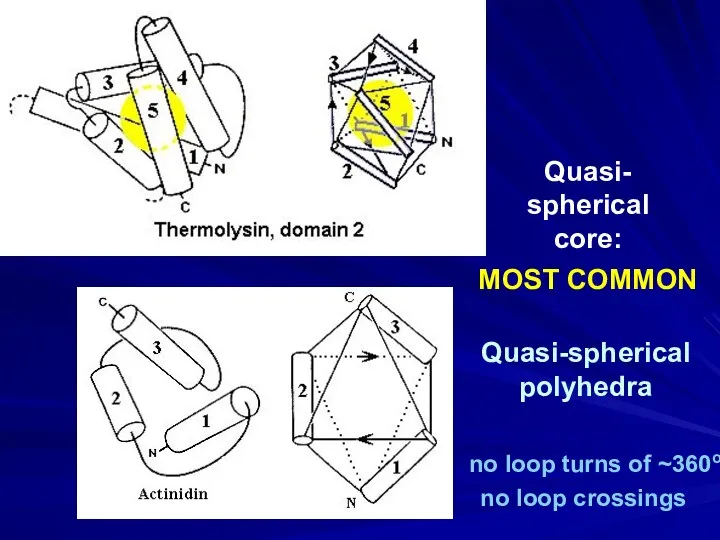
- 21. Quasi-spherical polyhedra Quasi- spherical core: MOST COMMON no loop turns of ~360o no loop crossings
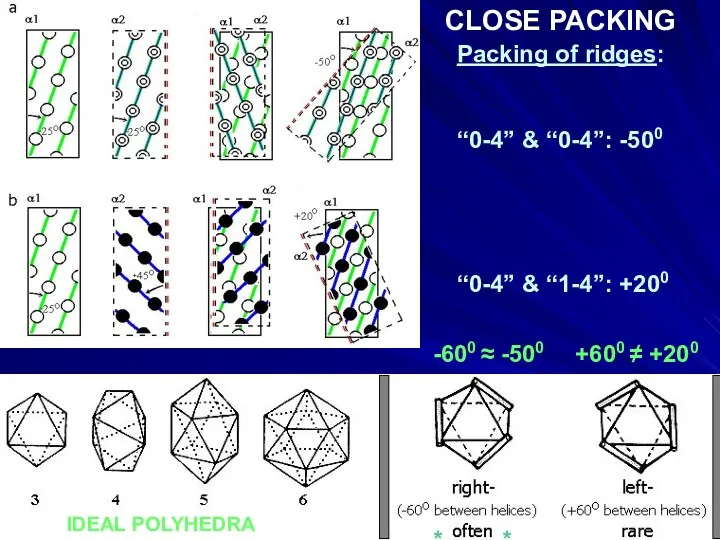
- 22. Packing of ridges: “0-4” & “0-4”: -500 “0-4” & “1-4”: +200 IDEAL POLYHEDRA -600 ≈ -500
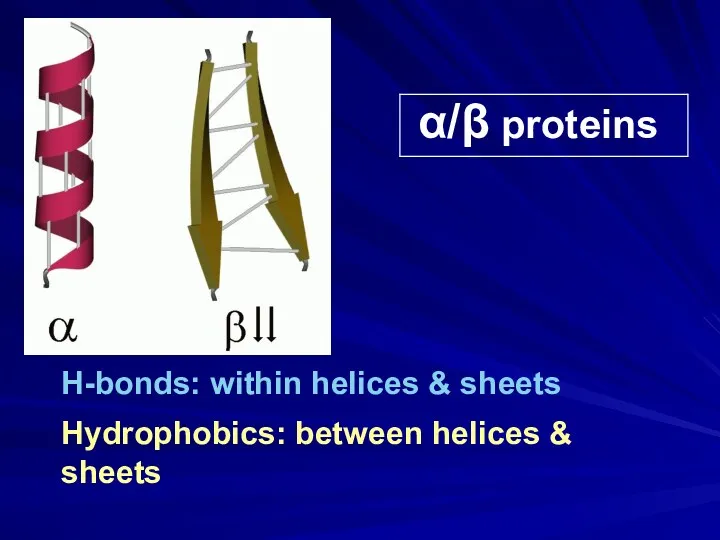
- 23. α/β proteins H-bonds: within helices & sheets Hydrophobics: between helices & sheets
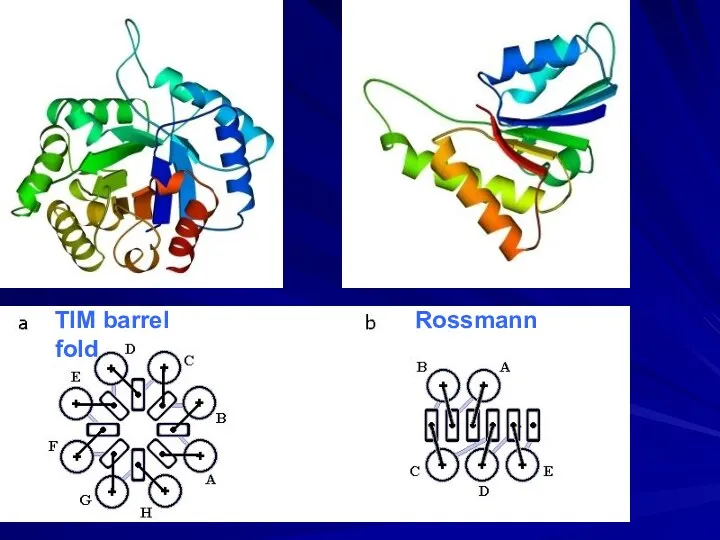
- 24. TIM barrel Rossmann fold
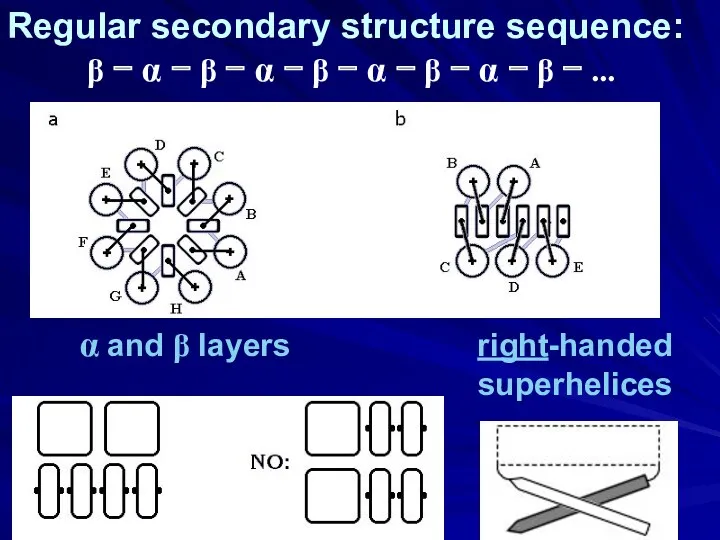
- 25. α and β layers right-handed superhelices Regular secondary structure sequence: β − α − β −
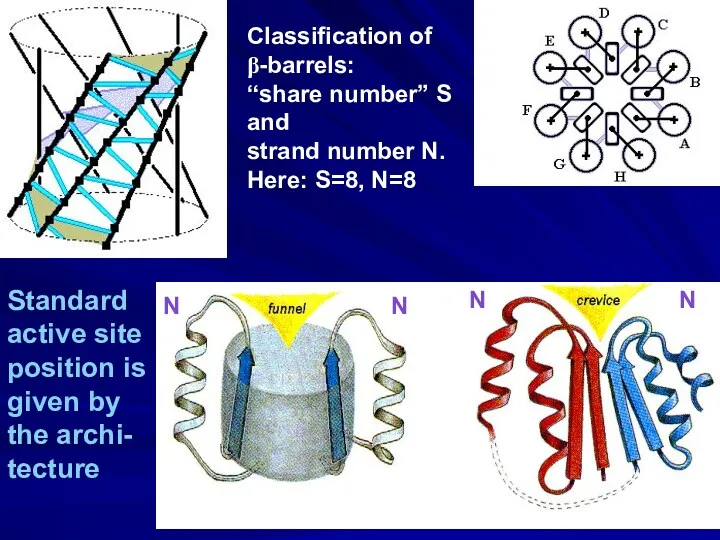
- 26. Classification of β-barrels: “share number” S and strand number N. Here: S=8, N=8 Standard active site
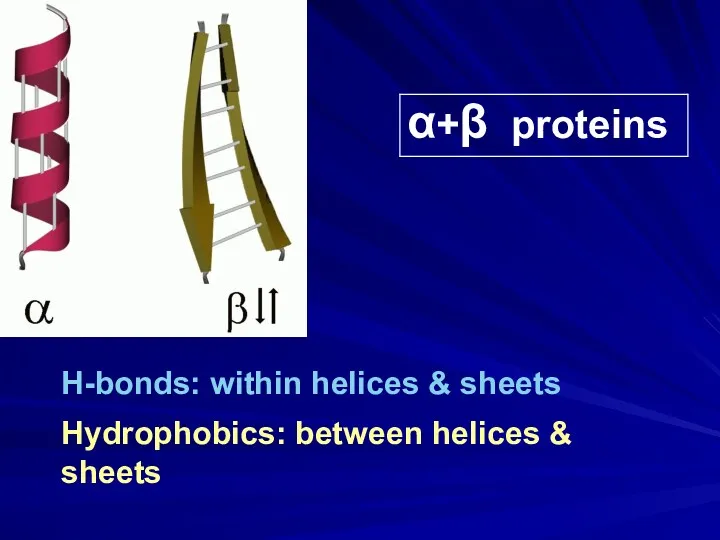
- 27. α+β proteins H-bonds: within helices & sheets Hydrophobics: between helices & sheets
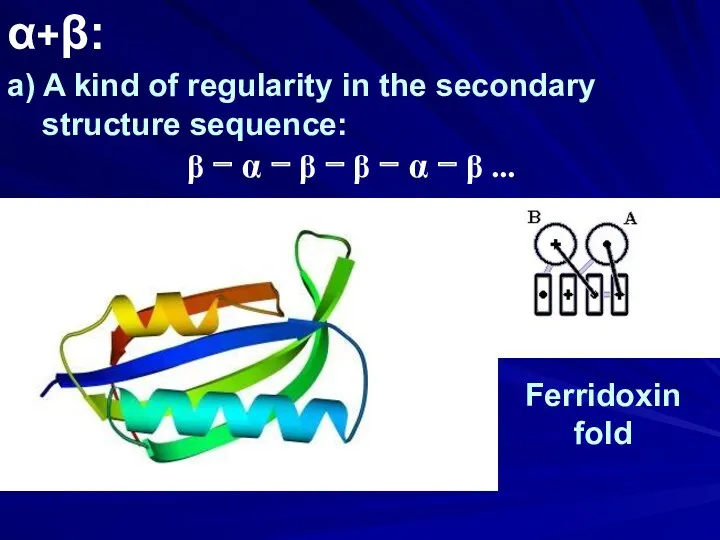
- 28. α+β: a) A kind of regularity in the secondary structure sequence: β − α − β
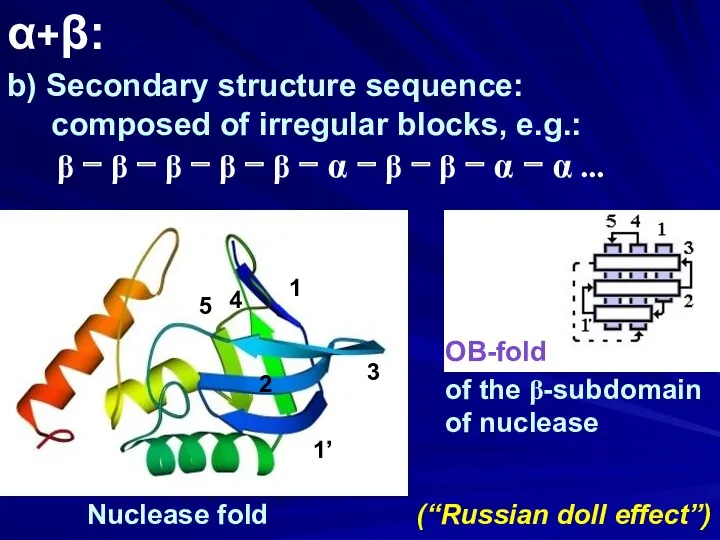
- 29. α+β: b) Secondary structure sequence: composed of irregular blocks, e.g.: β − β − β −
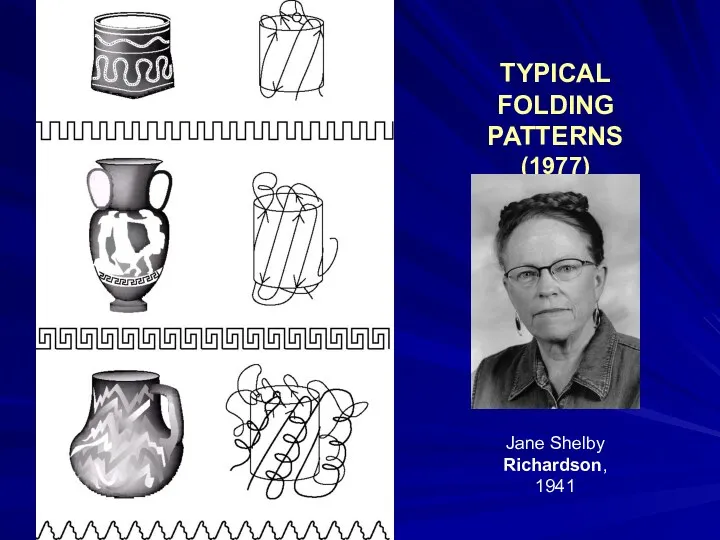
- 30. TYPICAL FOLDING PATTERNS (1977) Jane Shelby Richardson, 1941
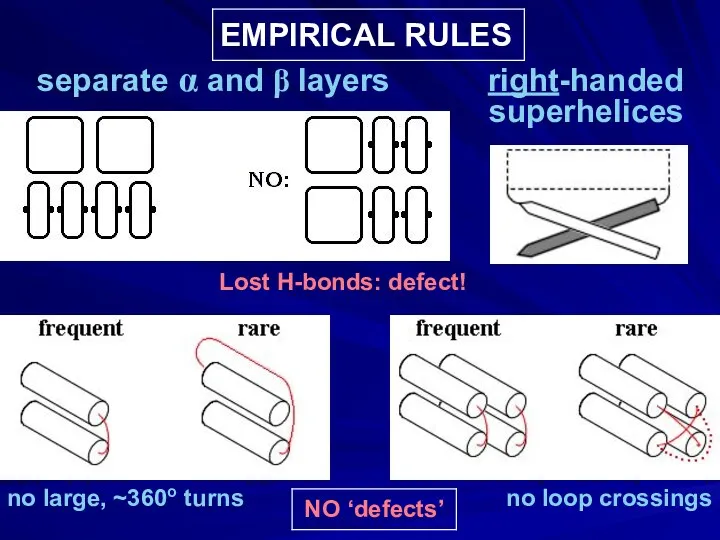
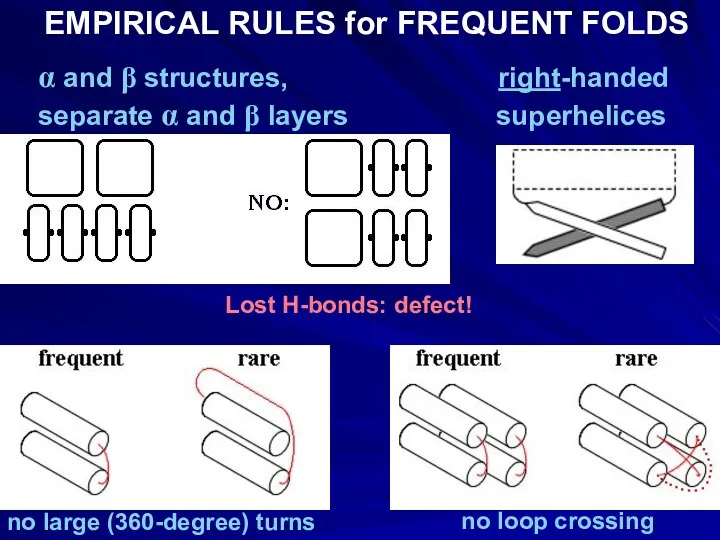
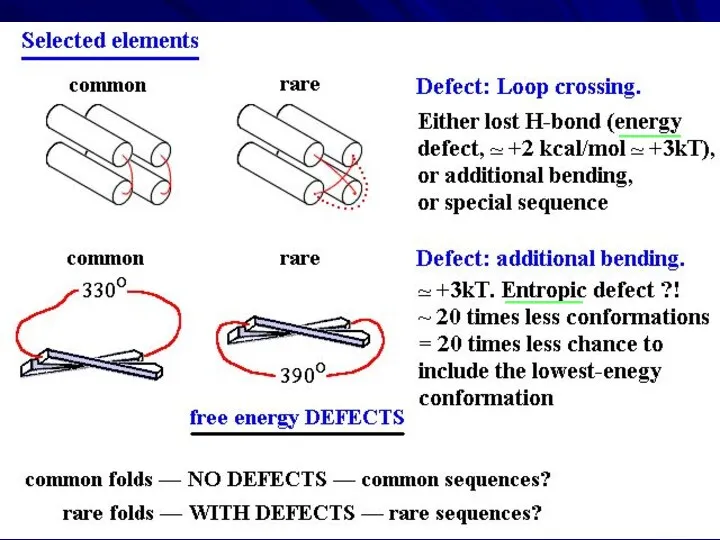
- 31. EMPIRICAL RULES separate α and β layers right-handed superhelices no large, ~360o turns no loop crossings
- 32. RESULT: NARROW SET OF PREDOMINANT FOLDING PATTERNS these are those that have no ‘defects’
- 33. ALSO, these are “natively disordered proteins”, which form a definite structure only when bound to some
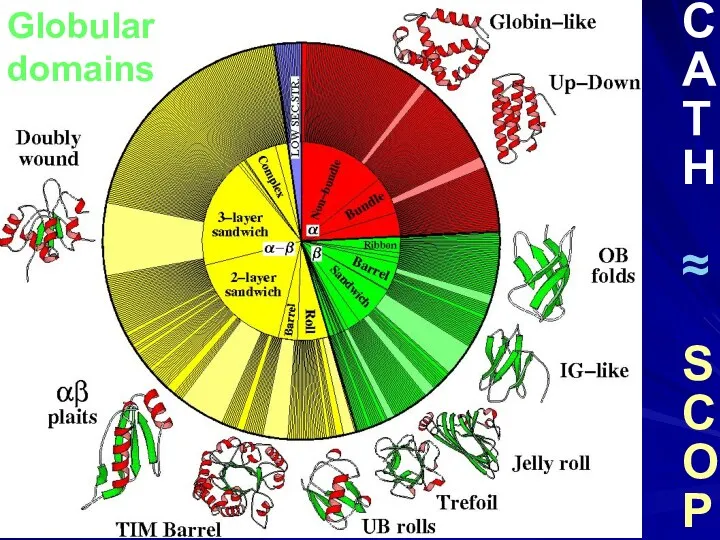
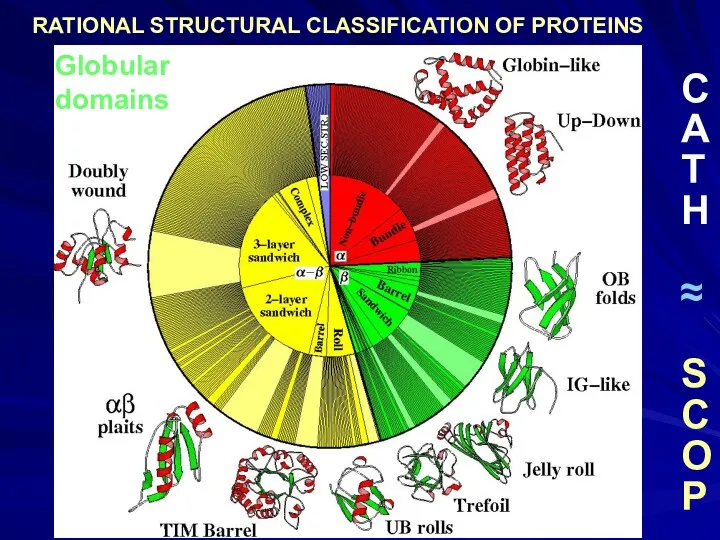
- 34. Globular domains C A T H ≈ S C O P
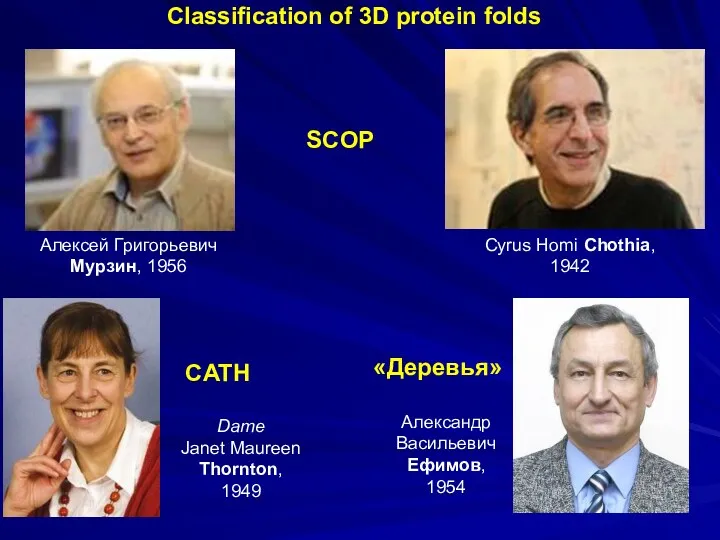
- 35. Алексей Григорьевич Мурзин, 1956 Dame Janet Maureen Thornton, 1949 Cyrus Homi Chothia, 1942 CATH SCOP Classification
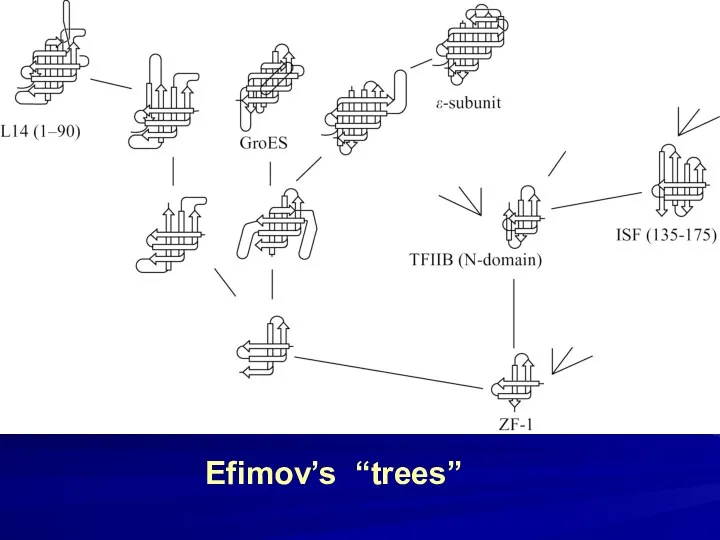
- 36. Efimov’s “trees”
- 37. 80/20 LAW:
- 38. EMPIRICAL RULES for FREQUENT FOLDS α and β structures, right-handed separate α and β layers superhelices
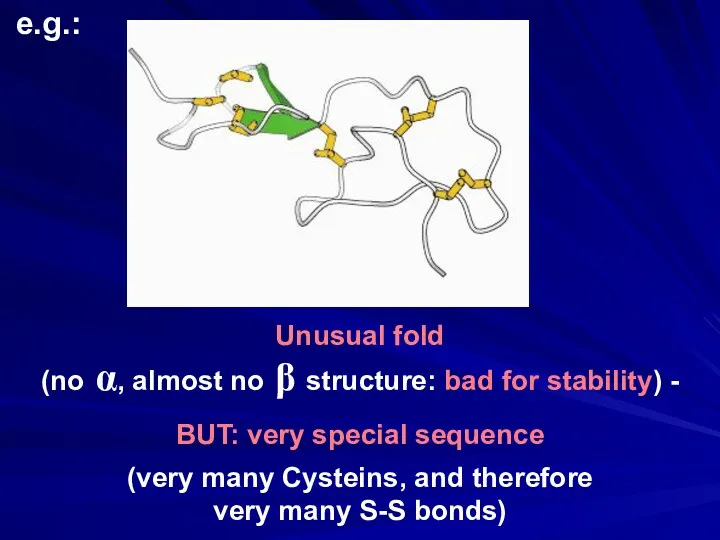
- 39. Unusual fold (no α, almost no β structure: bad for stability) - BUT: very special sequence
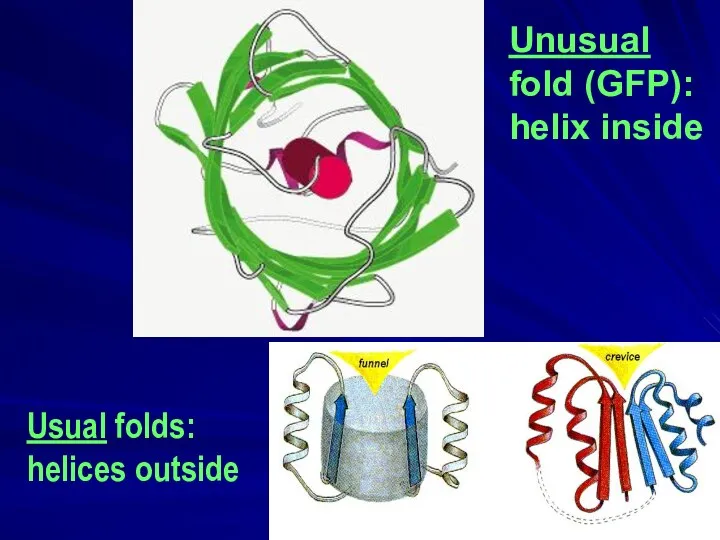
- 40. Unusual fold (GFP): helix inside Usual folds: helices outside
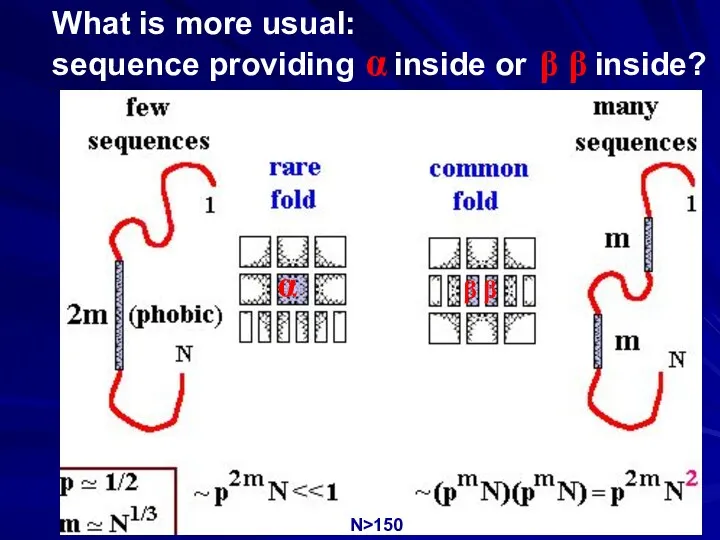
- 41. What is more usual: sequence providing α inside or β β inside? α β β N>150
- 42. _____ ____
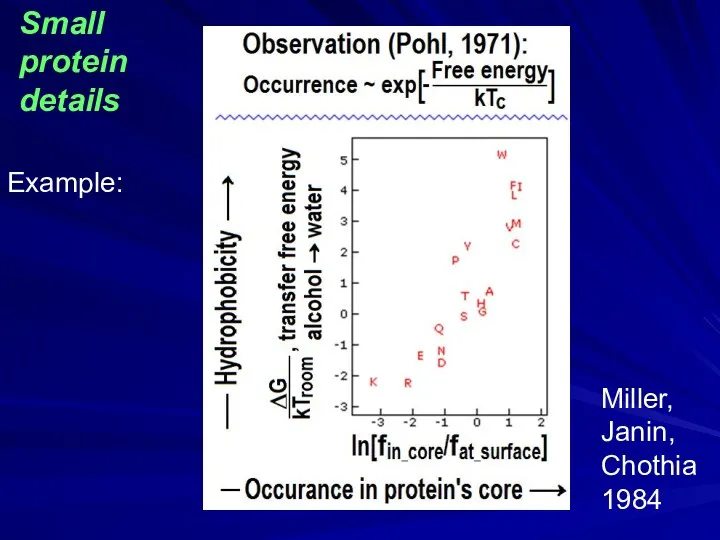
- 43. Miller, Janin, Chothia 1984 Example: Small protein details
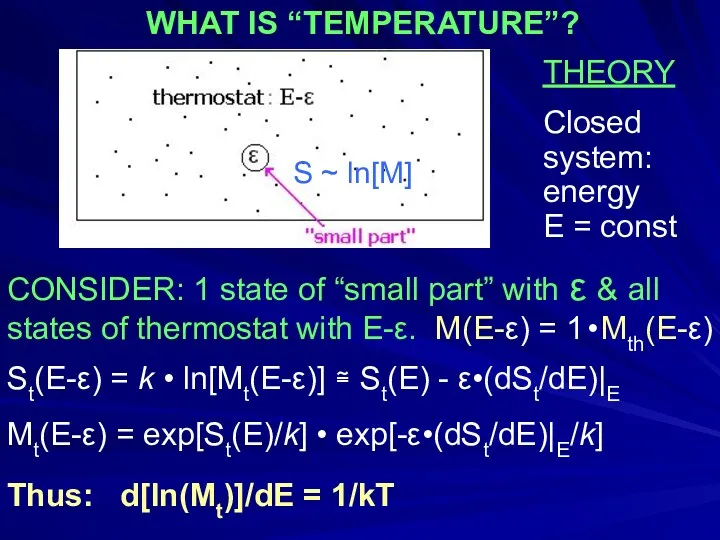
- 44. THEORY Closed system: energy E = const CONSIDER: 1 state of “small part” with ε &
- 45. Protein structure is stable, if its free energy is below some threshold For example: below that
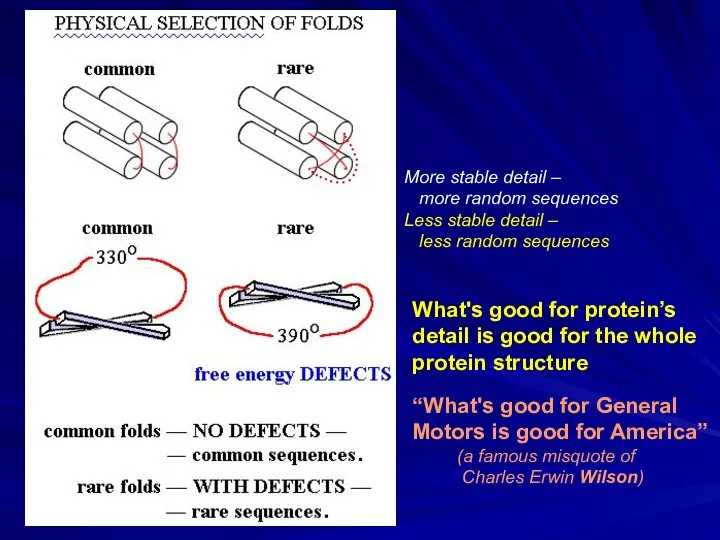
- 46. More stable detail – more random sequences Less stable detail – less random sequences What's good
- 47. “Multitude principle” for physical selection of folds of globular proteins (now: “designability”): the more sequences fit
- 48. Globular domains C A T H ≈ S C O P RATIONAL STRUCTURAL CLASSIFICATION OF PROTEINS
- 50. Скачать презентацию







 Интерференция света. (Лекция 11)
Интерференция света. (Лекция 11) Происхождение элементов
Происхождение элементов Кроссворд
Кроссворд Молекулярно-кинетическая теория строения вещества (МКТ)
Молекулярно-кинетическая теория строения вещества (МКТ) Кинематика тела. Лекция 7
Кинематика тела. Лекция 7 тест по состояниям вещества 8 класс
тест по состояниям вещества 8 класс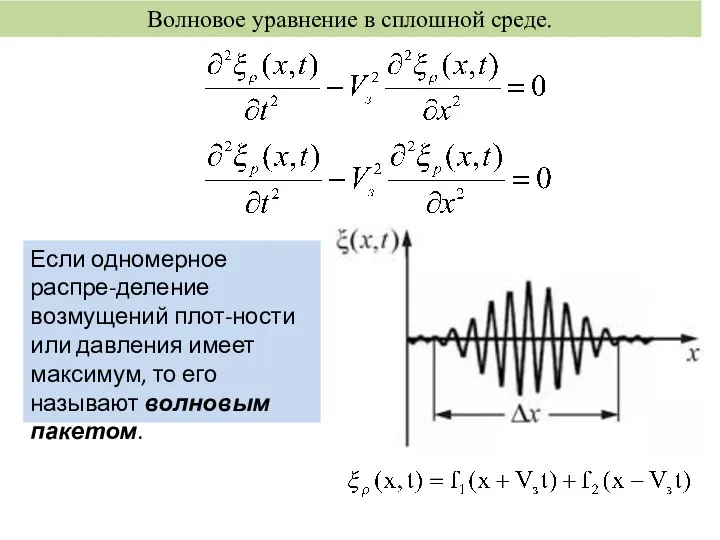 Волновое уравнение в сплошной среде
Волновое уравнение в сплошной среде Электромагнитные волны
Электромагнитные волны Методы исследования наносистем и наноматериалов. Классификация физико-химических методов исследования
Методы исследования наносистем и наноматериалов. Классификация физико-химических методов исследования Презентация к уроку физики в 8 классе: Обобщение по теме Агрегатные состояния вещества.
Презентация к уроку физики в 8 классе: Обобщение по теме Агрегатные состояния вещества. Магнитные материалы. Магнитное поле в веществе
Магнитные материалы. Магнитное поле в веществе Урок физики 8 класс Испарение и конденсация
Урок физики 8 класс Испарение и конденсация Аеродинамічні характеристики літака. Аеродинамічне компонування літака. (Лекція 7.4.1)
Аеродинамічні характеристики літака. Аеродинамічне компонування літака. (Лекція 7.4.1) 03.21г. Типы спектров. Спектральный анализ
03.21г. Типы спектров. Спектральный анализ Фундаментальные взаимодействия
Фундаментальные взаимодействия Презентация к уроку Водород, 5 класс, предмет Физика и химия
Презентация к уроку Водород, 5 класс, предмет Физика и химия Закон Гука
Закон Гука Лабораторная работа №2. Измерение массы тела на рычажных весах
Лабораторная работа №2. Измерение массы тела на рычажных весах Электризация тел. Два рода зарядов
Электризация тел. Два рода зарядов Поляризация поперечных волн. Поляризация света. Дисперсия света. Виды спектров
Поляризация поперечных волн. Поляризация света. Дисперсия света. Виды спектров Батарейка своими руками
Батарейка своими руками Качество электроэнергии
Качество электроэнергии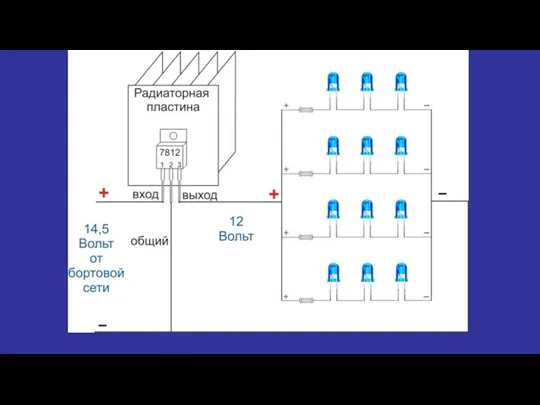 Электрическая схема светильника с регулировкой мощности
Электрическая схема светильника с регулировкой мощности Шлифовальные станки. (Тема 7)
Шлифовальные станки. (Тема 7) Электромагнитная картина мира
Электромагнитная картина мира Курсовая работа по теоретической механике “Динамика кулисного механизма”
Курсовая работа по теоретической механике “Динамика кулисного механизма”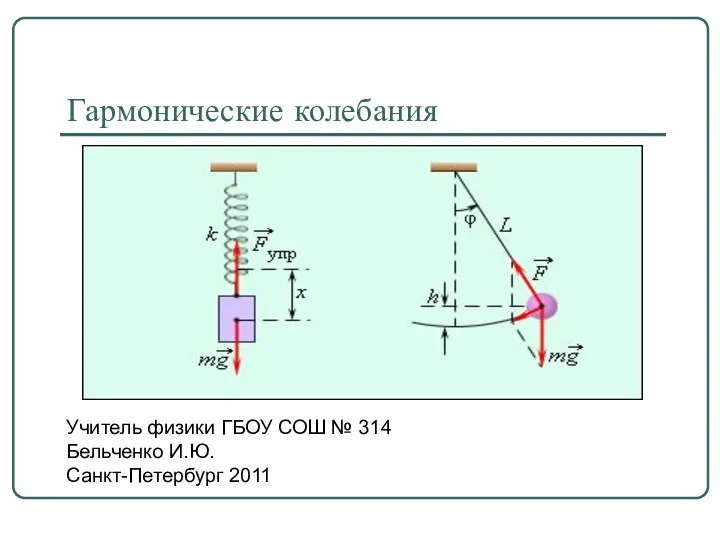 Гармонические колебания
Гармонические колебания Работа силы Ампера. Магнитный поток. Явление электромагнитной индукции. Закон электромагнитной индукции. Правило Ленца
Работа силы Ампера. Магнитный поток. Явление электромагнитной индукции. Закон электромагнитной индукции. Правило Ленца