Содержание
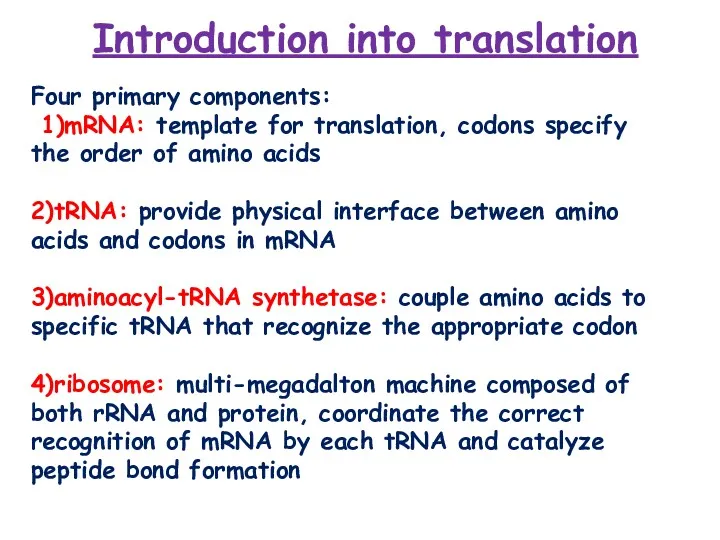
- 2. Introduction into translation Four primary components: 1)mRNA: template for translation, codons specify the order of amino
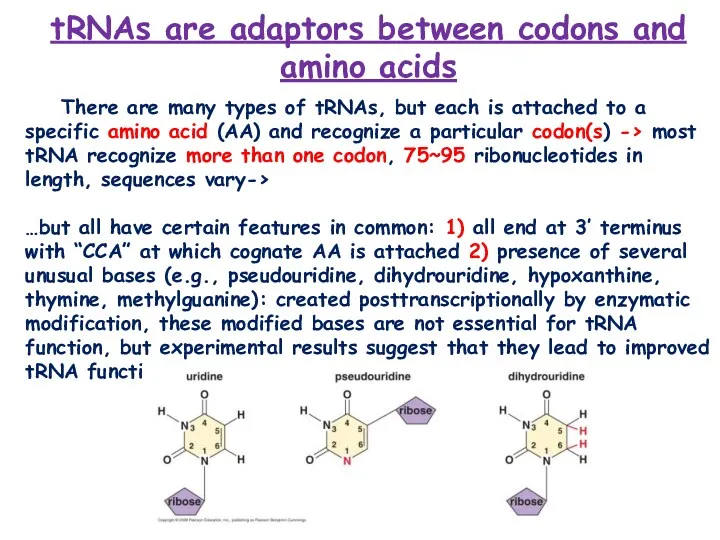
- 3. tRNAs are adaptors between codons and amino acids There are many types of tRNAs, but each
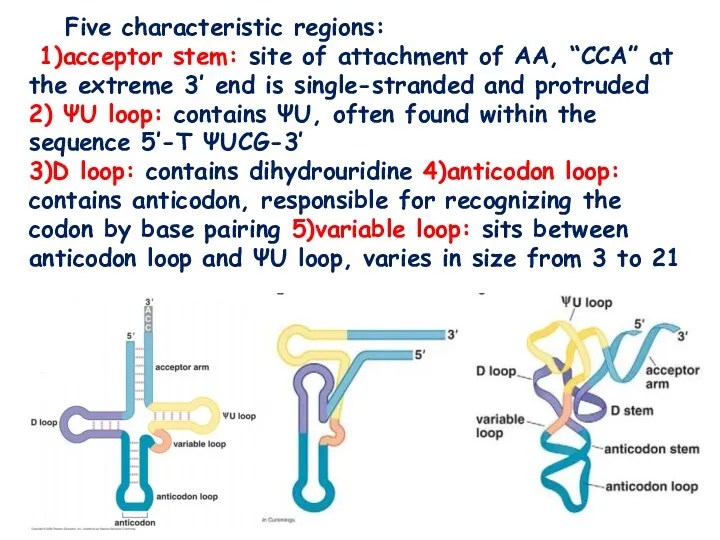
- 4. Five characteristic regions: 1)acceptor stem: site of attachment of AA, “CCA” at the extreme 3’ end
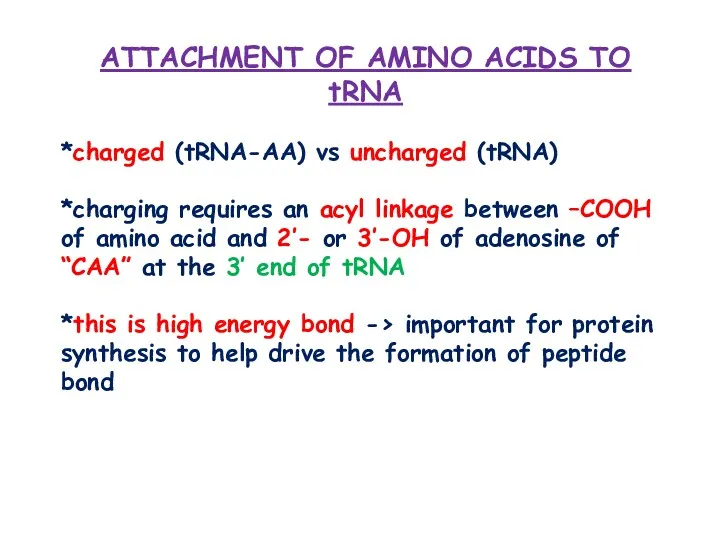
- 5. ATTACHMENT OF AMINO ACIDS TO tRNA *charged (tRNA-AA) vs uncharged (tRNA) *charging requires an acyl linkage
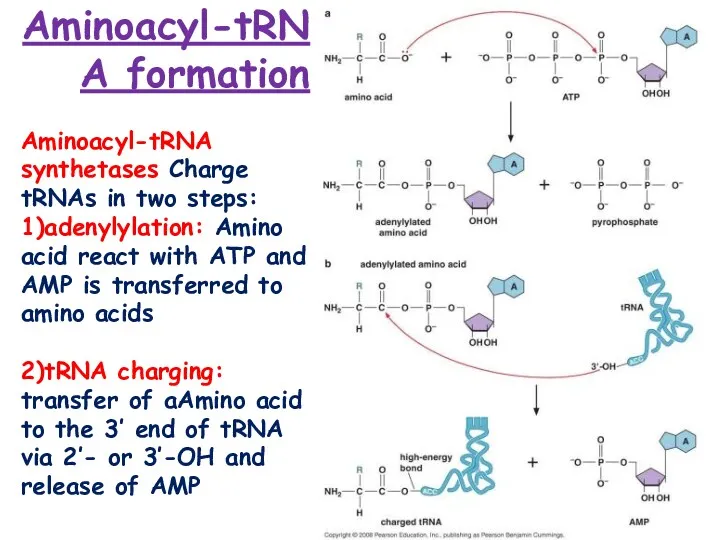
- 6. Aminoacyl-tRNA formation Aminoacyl-tRNA synthetases Charge tRNAs in two steps: 1)adenylylation: Amino acid react with ATP and
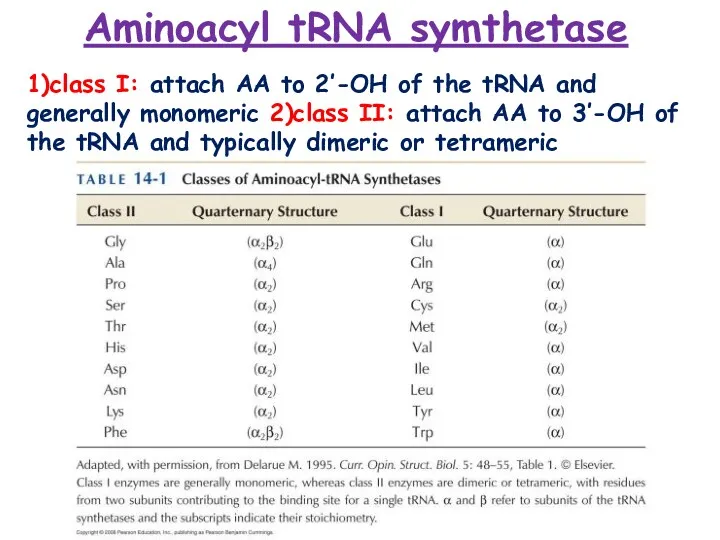
- 7. Aminoacyl tRNA symthetase 1)class I: attach AA to 2’-OH of the tRNA and generally monomeric 2)class
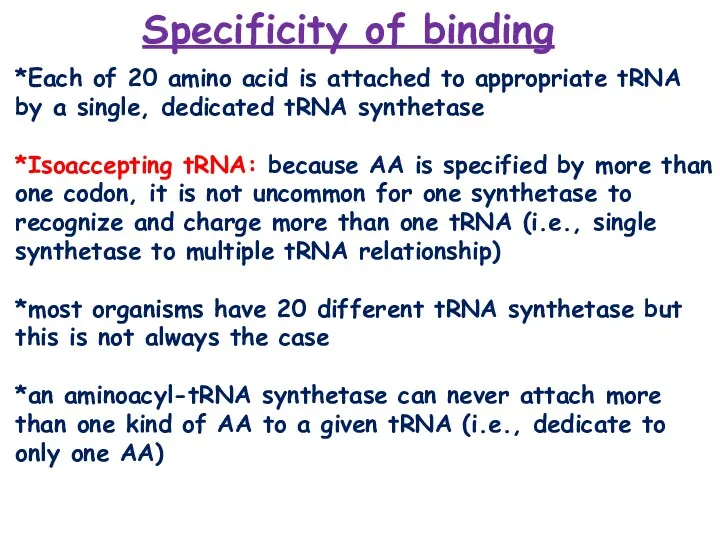
- 8. *Each of 20 amino acid is attached to appropriate tRNA by a single, dedicated tRNA synthetase
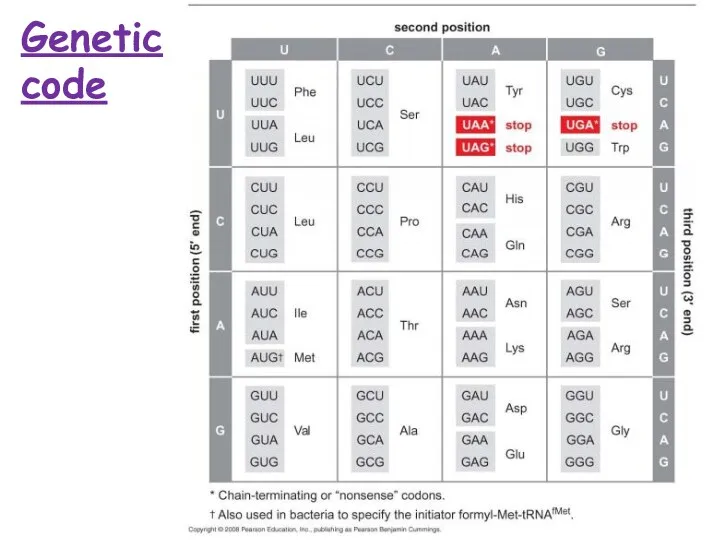
- 9. Genetic code
- 10. What features of tRNA enable synthetase to discriminate the correct set of tRNA from tRNA for
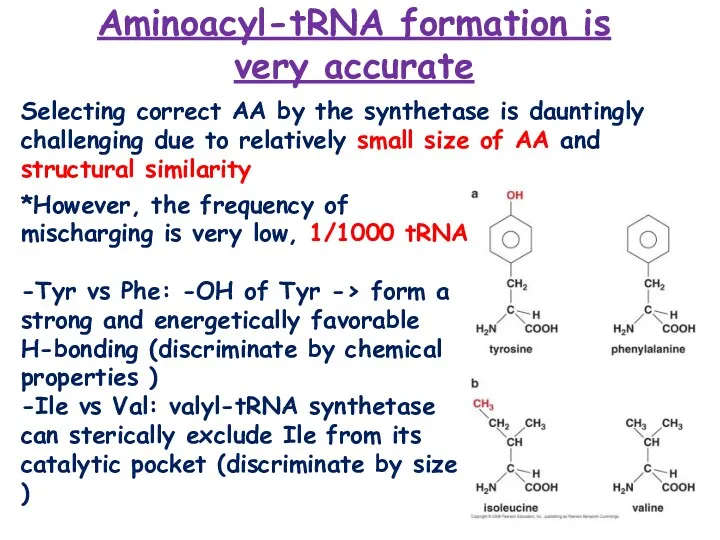
- 11. Aminoacyl-tRNA formation is very accurate Selecting correct AA by the synthetase is dauntingly challenging due to
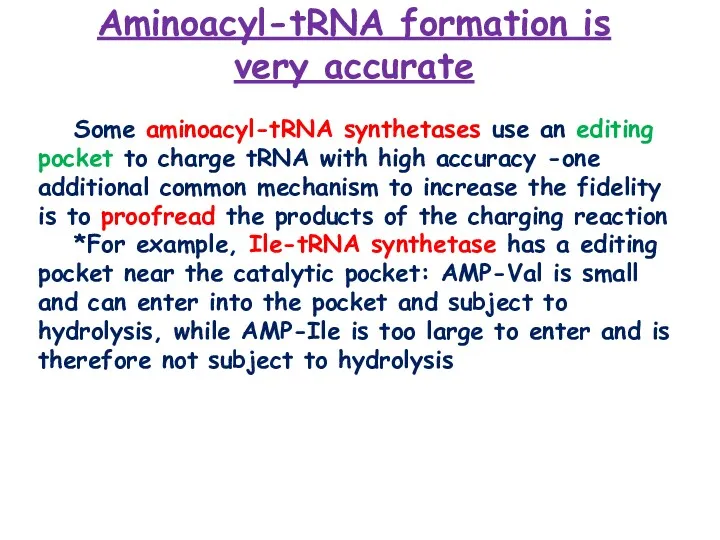
- 12. Some aminoacyl-tRNA synthetases use an editing pocket to charge tRNA with high accuracy -one additional common
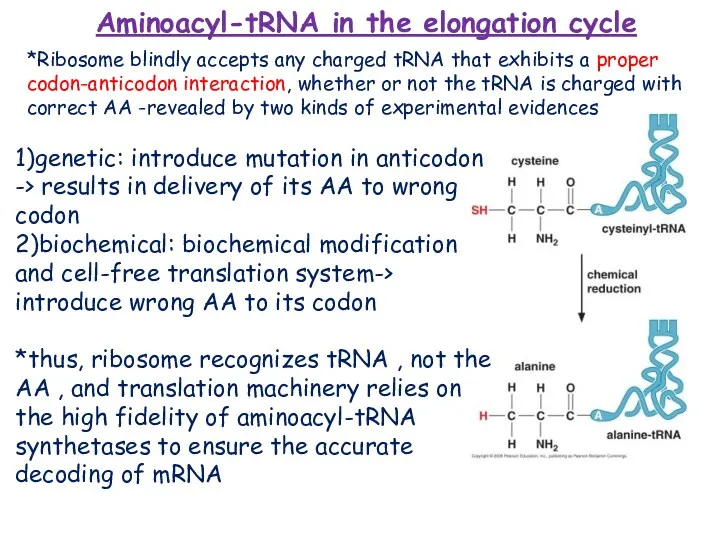
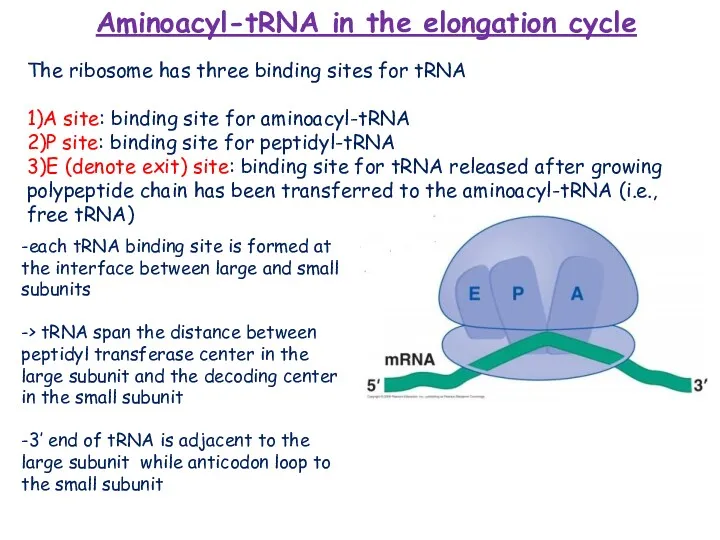
- 13. Aminoacyl-tRNA in the elongation cycle 1)genetic: introduce mutation in anticodon -> results in delivery of its
- 14. -each tRNA binding site is formed at the interface between large and small subunits -> tRNA
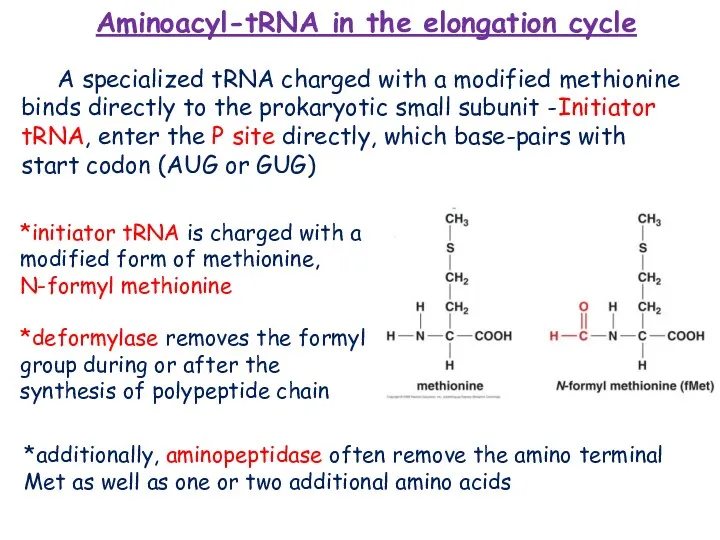
- 15. *initiator tRNA is charged with a modified form of methionine, N-formyl methionine *deformylase removes the formyl
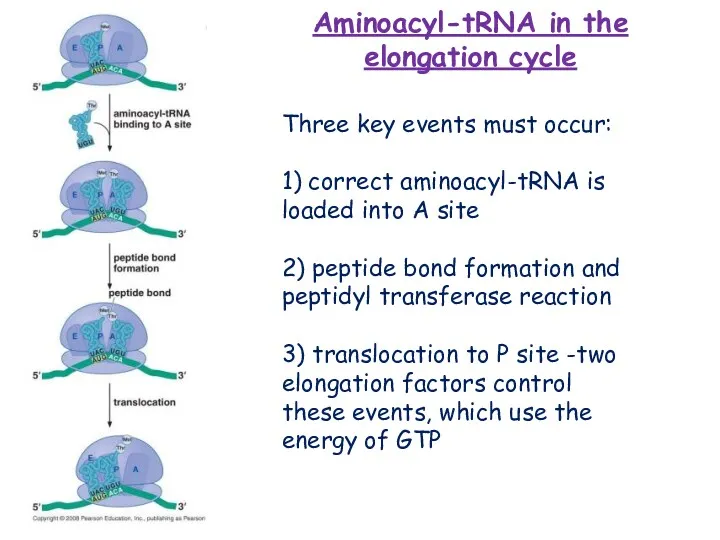
- 16. Three key events must occur: 1) correct aminoacyl-tRNA is loaded into A site 2) peptide bond
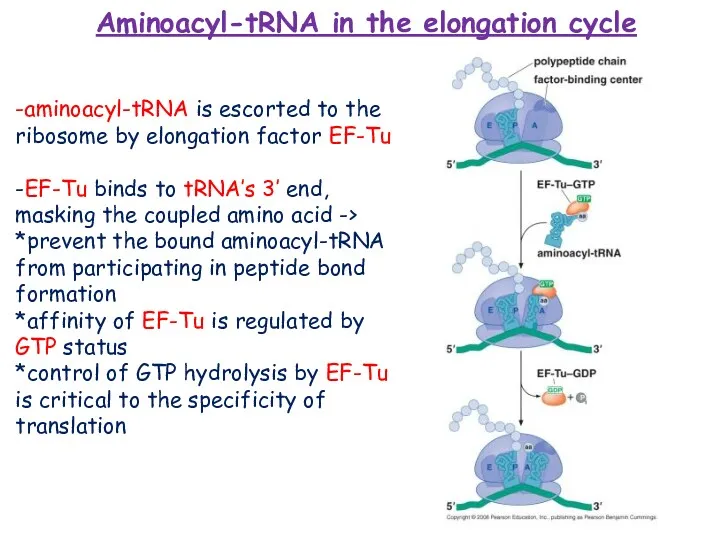
- 17. -aminoacyl-tRNA is escorted to the ribosome by elongation factor EF-Tu -EF-Tu binds to tRNA’s 3’ end,
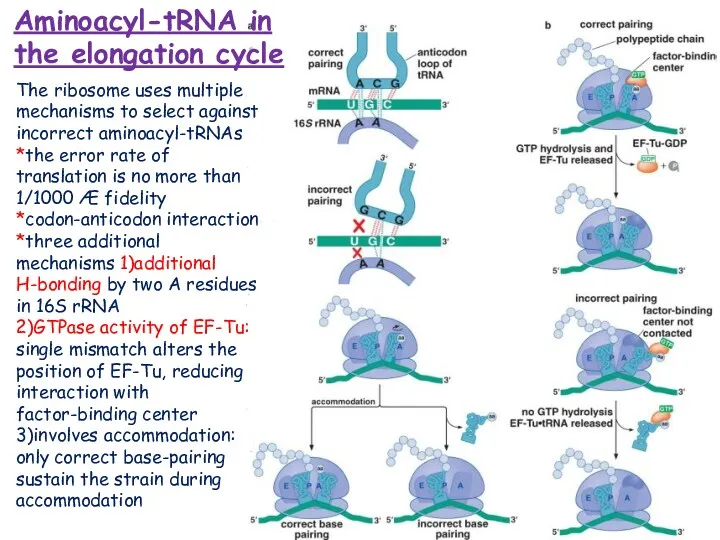
- 18. Aminoacyl-tRNA in the elongation cycle The ribosome uses multiple mechanisms to select against incorrect aminoacyl-tRNAs *the
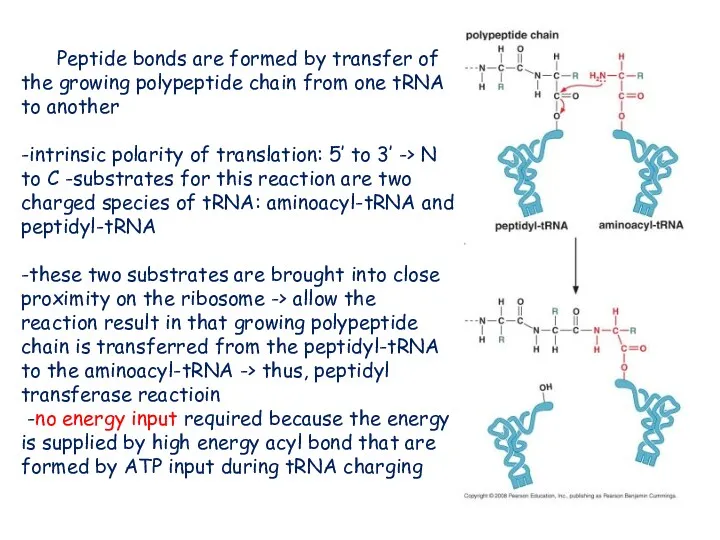
- 19. Peptide bonds are formed by transfer of the growing polypeptide chain from one tRNA to another
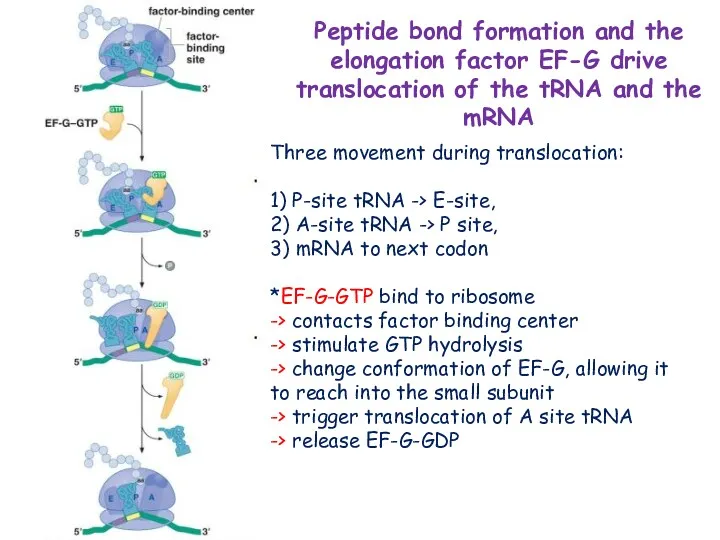
- 20. Three movement during translocation: 1) P-site tRNA -> E-site, 2) A-site tRNA -> P site, 3)
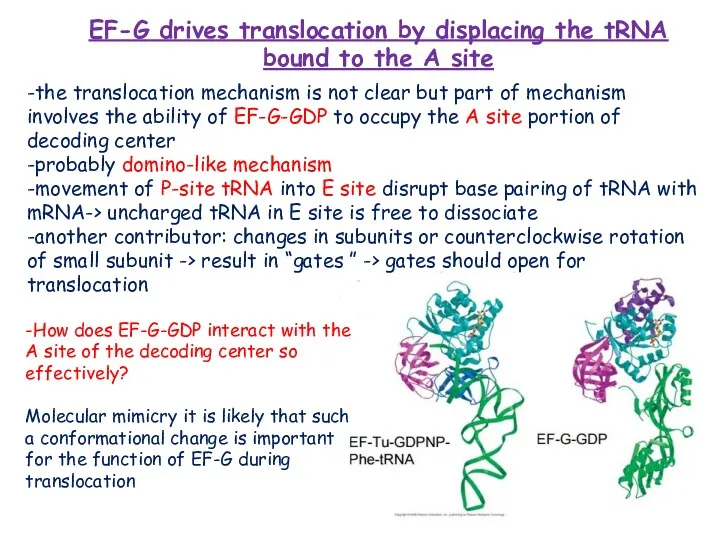
- 21. EF-G drives translocation by displacing the tRNA bound to the A site -the translocation mechanism is
- 22. During protein synthesis dictated by the codon sequence of messenger RNA, the ribosome selects aminoacyltRNA (aa-tRNA)
- 23. REFERENCE Watson J D, Baker T A , Bell S P, Gann A, Levine M, Losick
- 25. Скачать презентацию

 Environmental pollution
Environmental pollution Salty dough products
Salty dough products Days of the week
Days of the week Travelling
Travelling Uncountable Nouns
Uncountable Nouns We count that lake Baikal need protect
We count that lake Baikal need protect Past Progressive tense. Употребление времени
Past Progressive tense. Употребление времени Английские цифры от 1 до 10
Английские цифры от 1 до 10 Hobby in Britain
Hobby in Britain Having Fun!
Having Fun! The nature, types and functions of Lexical Stylistic Devices: Irony
The nature, types and functions of Lexical Stylistic Devices: Irony Modal verbs
Modal verbs Private school in UK
Private school in UK Riddles about animals. Загадки о животных. (2 класс)
Riddles about animals. Загадки о животных. (2 класс) Canada is located in the north of the American
Canada is located in the north of the American Modal verbs. Значение, использование, эквиваленты
Modal verbs. Значение, использование, эквиваленты What are they going to do?
What are they going to do? My English portfolio. Образец портфолио ученика на английском языке
My English portfolio. Образец портфолио ученика на английском языке Nonfiction. Text. Features
Nonfiction. Text. Features Jeopardy. Quiz
Jeopardy. Quiz The subjunctive mood
The subjunctive mood I can. I can’t
I can. I can’t Colors
Colors Образование порядковых числительных
Образование порядковых числительных The definite article with geographical names
The definite article with geographical names Упражнение времена группы past
Упражнение времена группы past Guess these food idioms
Guess these food idioms Spotlight 3. Module 1 (Unit 1). School days
Spotlight 3. Module 1 (Unit 1). School days