Содержание
- 2. Comparing RNA and DNA DNA can replicate itself precisely and contain information in the specific sequence
- 3. Messenger RNA mRNA carries the message. The linear amino acid sequence (primary) is encoded in the
- 4. Basic Principles of Transcription and Translation RNA is the intermediate between genes and the proteins for
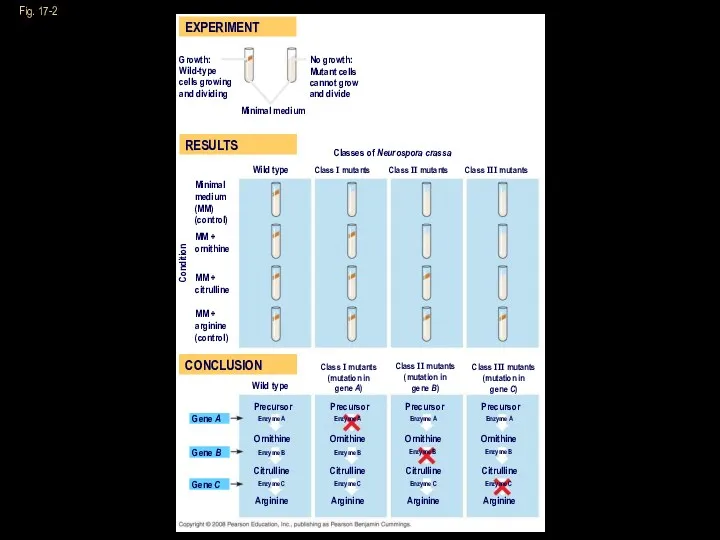
- 5. Fig. 17-2 RESULTS EXPERIMENT CONCLUSION Growth: Wild-type cells growing and dividing No growth: Mutant cells cannot
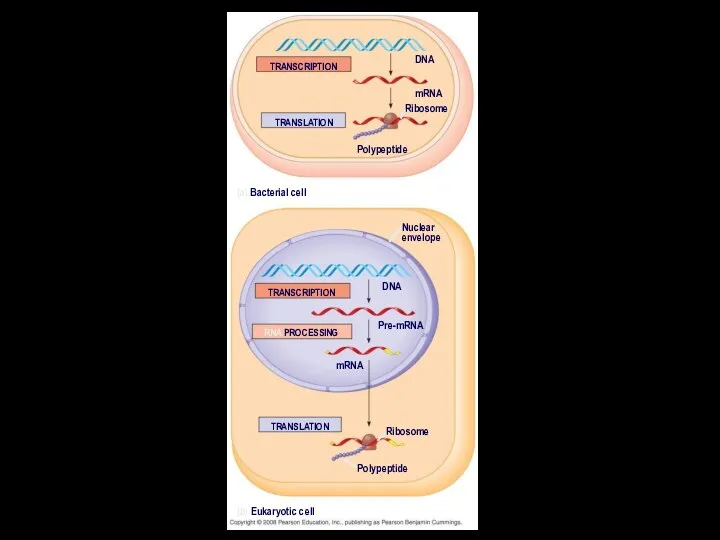
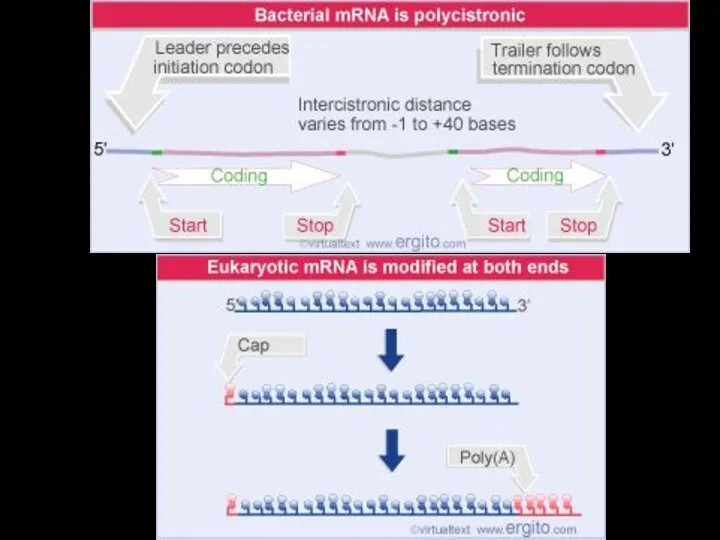
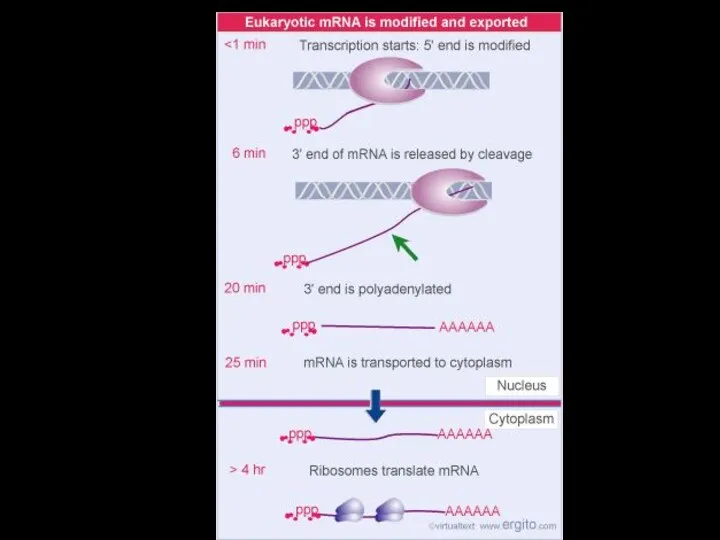
- 6. In prokaryotes, mRNA produced by transcription is immediately translated without more processing In a eukaryotic cell,
- 7. TRANSCRIPTION TRANSLATION DNA mRNA Ribosome Polypeptide (a) Bacterial cell Nuclear envelope TRANSCRIPTION RNA PROCESSING Pre-mRNA DNA
- 8. The Genetic Code The genetic code is the same for all organisms (universal) A codon is
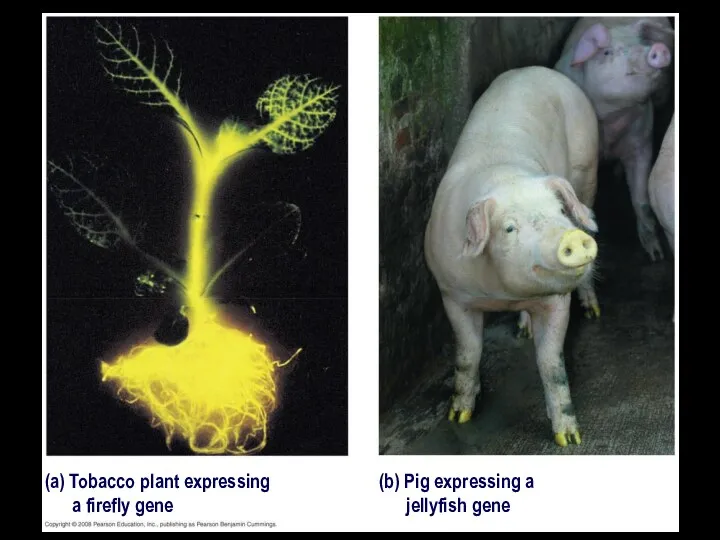
- 9. (a) Tobacco plant expressing a firefly gene (b) Pig expressing a jellyfish gene
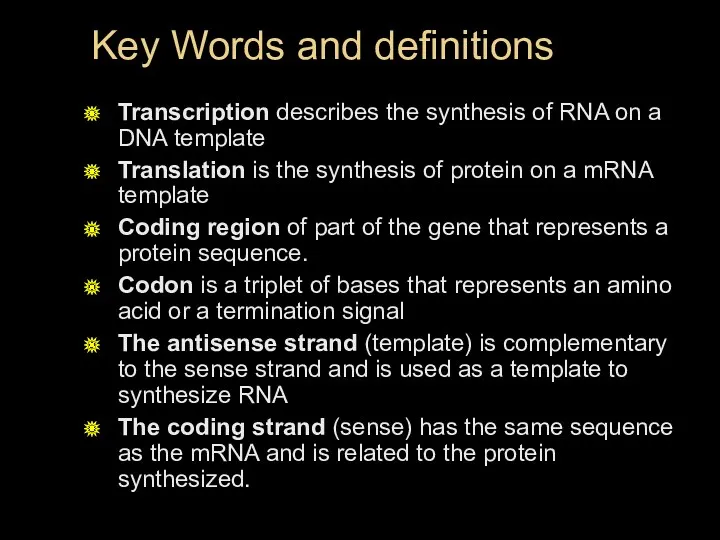
- 10. Key Words and definitions Transcription describes the synthesis of RNA on a DNA template Translation is
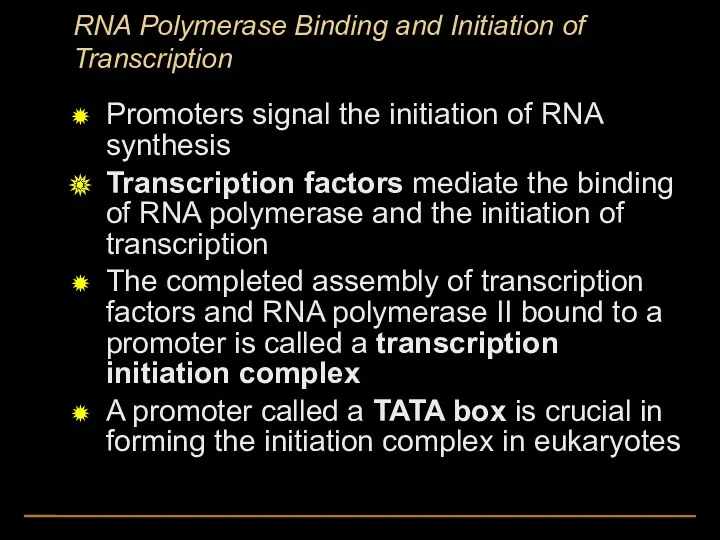
- 11. RNA Polymerase Binding and Initiation of Transcription Promoters signal the initiation of RNA synthesis Transcription factors
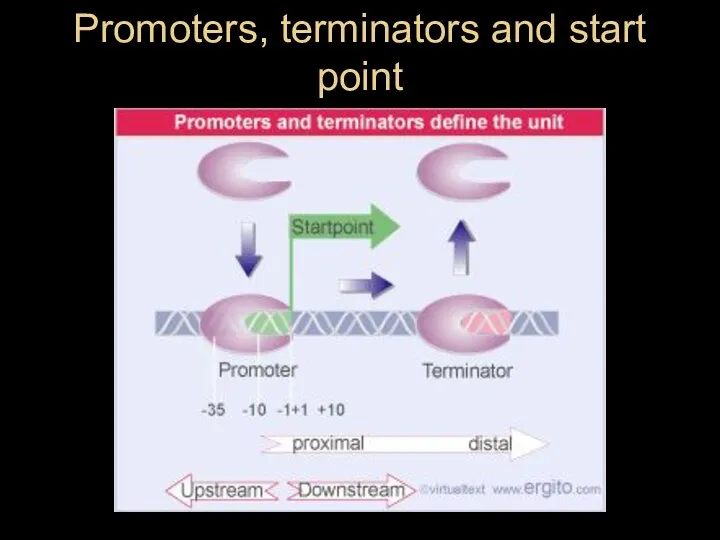
- 12. Promoters, terminators and start point
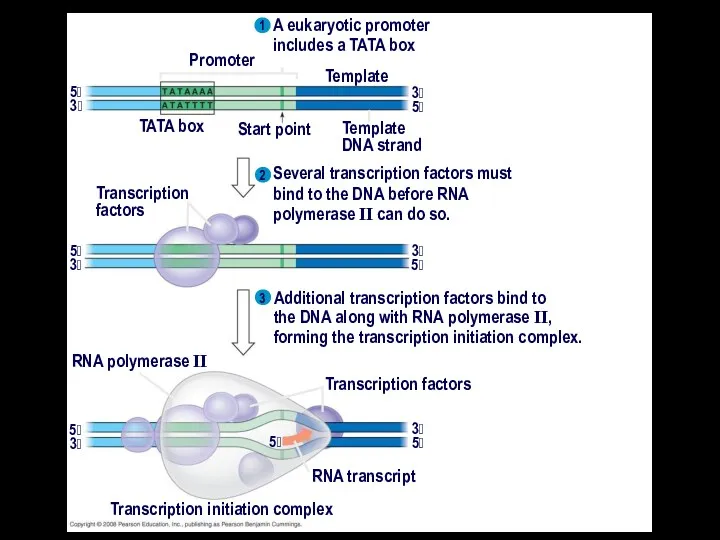
- 13. A eukaryotic promoter includes a TATA box 3 1 2 3 Promoter TATA box Start point
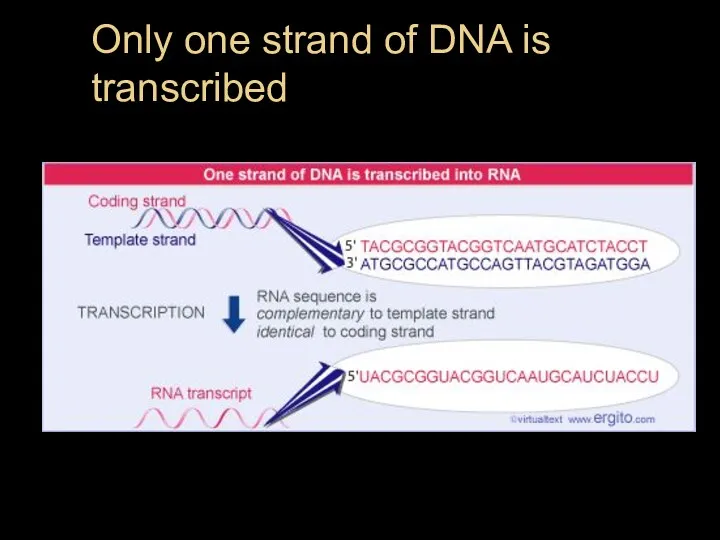
- 14. Only one strand of DNA is transcribed
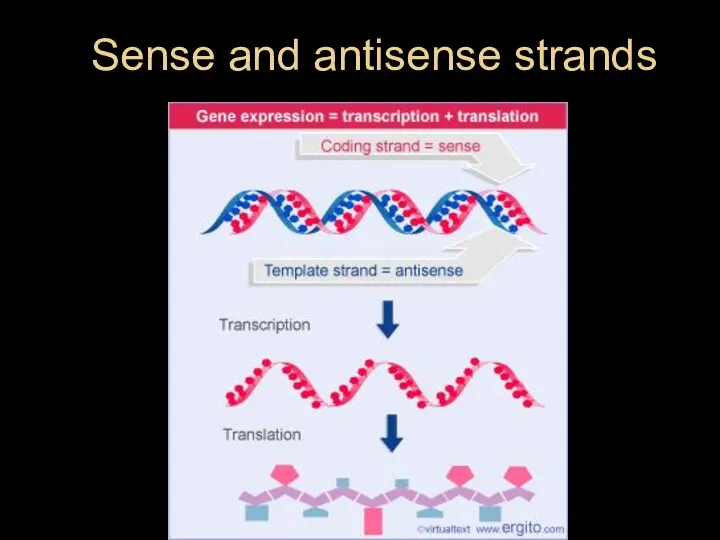
- 15. Sense and antisense strands
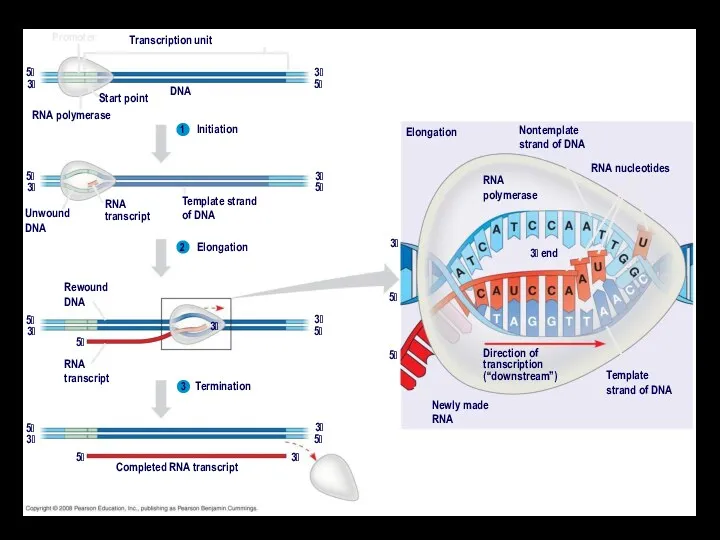
- 17. Promoter Transcription unit Start point DNA RNA polymerase 5 5 3 3 Initiation 1 2 3
- 18. Protein-coding segment Polyadenylation signal 3 3 UTR 5 UTR 5 5 Cap Start codon Stop codon
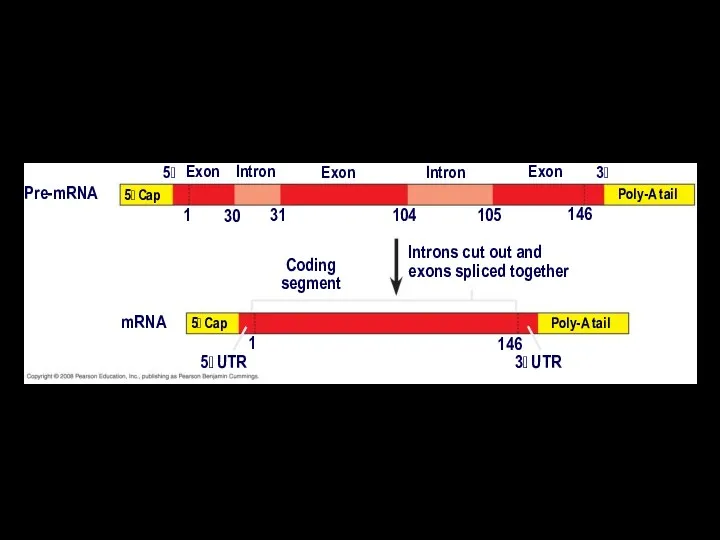
- 20. Split Genes and RNA Splicing Most eukaryotic genes and their RNA transcripts have long noncoding stretches
- 21. Pre-mRNA mRNA Coding segment Introns cut out and exons spliced together 5 Cap Exon Intron 5
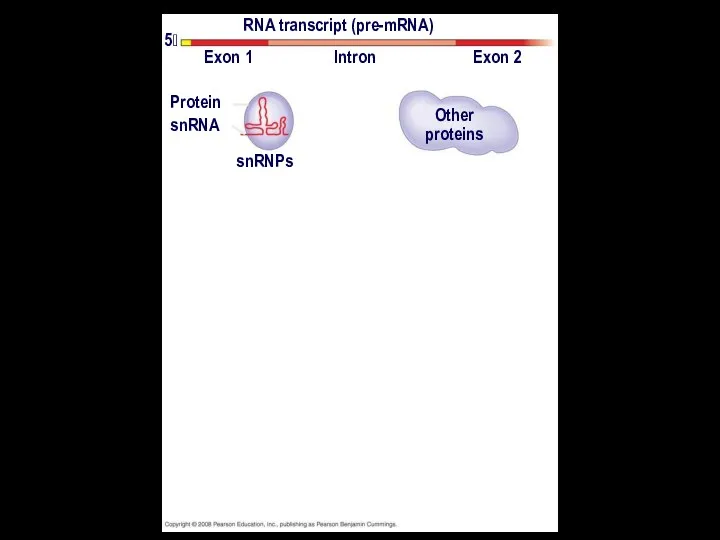
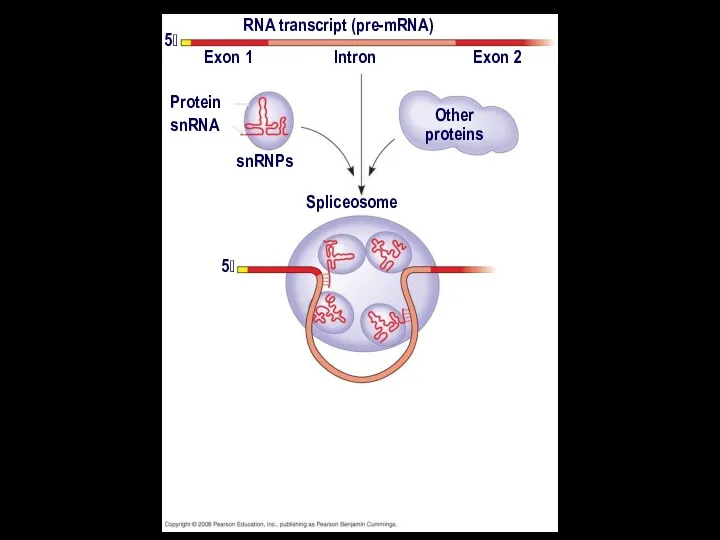
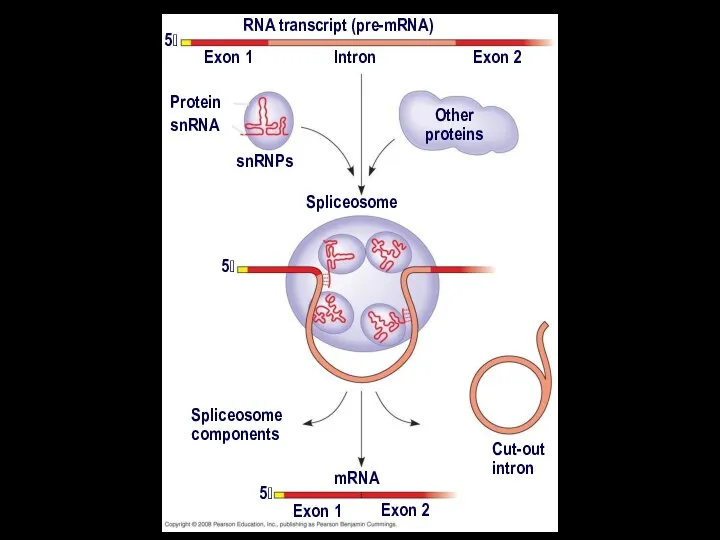
- 22. In some cases, RNA splicing is carried out by spliceosomes Spliceosomes consist of a variety of
- 23. RNA transcript (pre-mRNA) Exon 1 Exon 2 Intron Protein snRNA snRNPs Other proteins 5
- 24. RNA transcript (pre-mRNA) Exon 1 Exon 2 Intron Protein snRNA snRNPs Other proteins 5 5 Spliceosome
- 25. RNA transcript (pre-mRNA) Exon 1 Exon 2 Intron Protein snRNA snRNPs Other proteins 5 5 Spliceosome
- 26. Ribozymes Ribozymes are catalytic RNA molecules that function as enzymes and can splice RNA The discovery
- 27. Three properties of RNA enable it to function as an enzyme It can form a three-dimensional
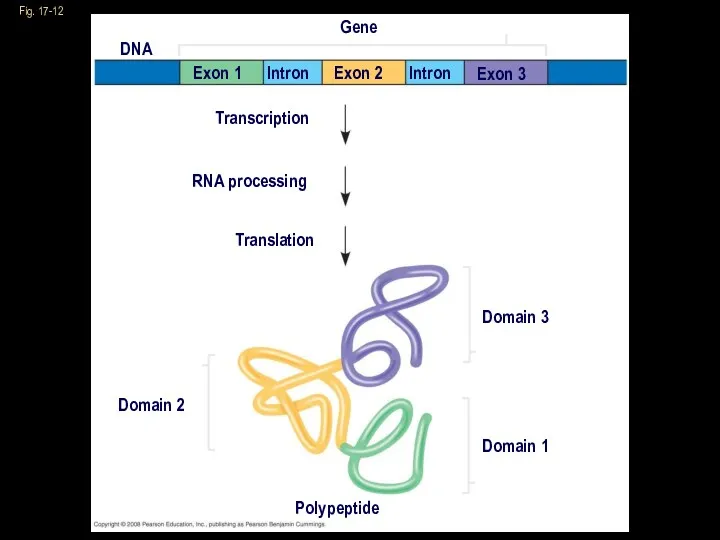
- 28. The Functional and Evolutionary Importance of Introns Some genes can encode more than one kind of
- 29. Proteins often have a modular architecture consisting of discrete regions called domains In many cases, different
- 30. Fig. 17-12 Gene DNA Exon 1 Exon 2 Exon 3 Intron Intron Transcription RNA processing Translation
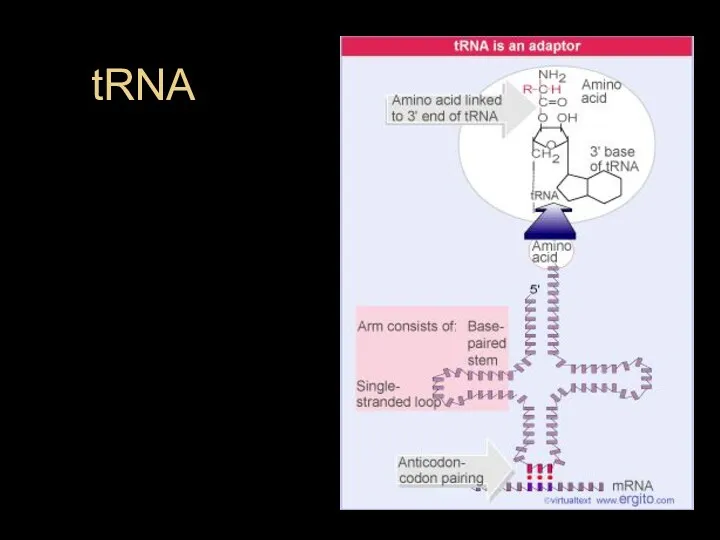
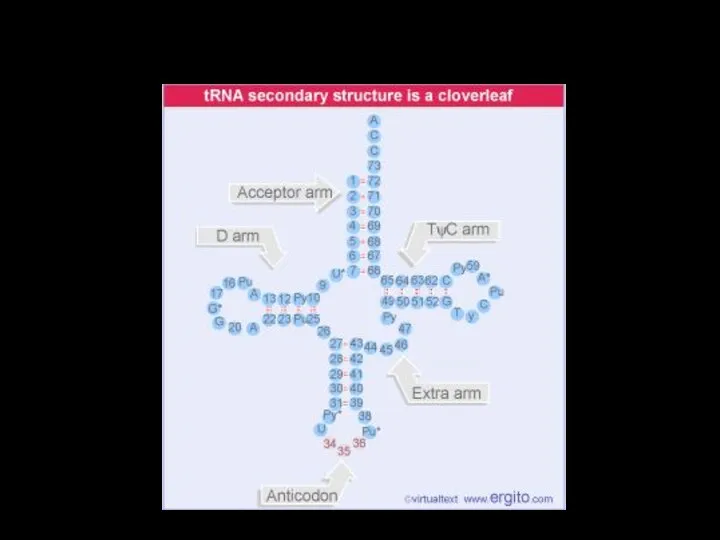
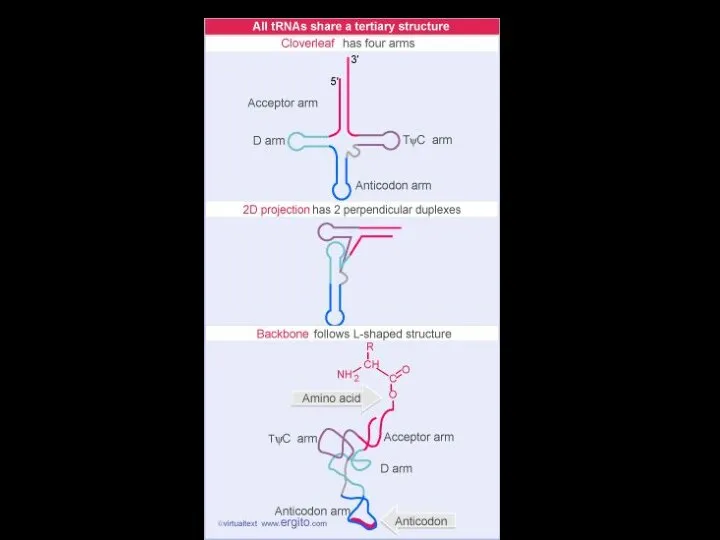
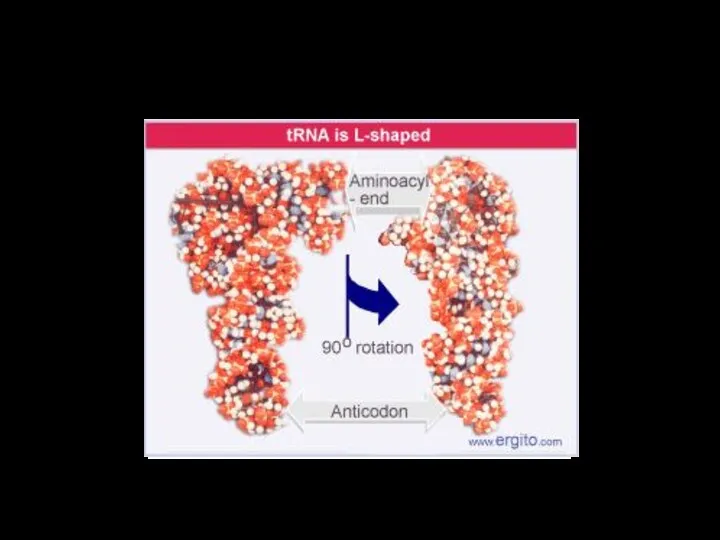
- 31. tRNA
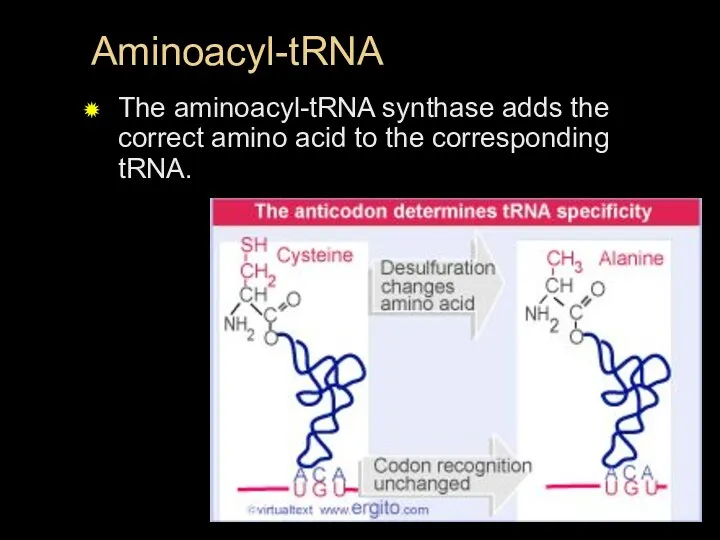
- 33. Aminoacyl-tRNA The aminoacyl-tRNA synthase adds the correct amino acid to the corresponding tRNA.
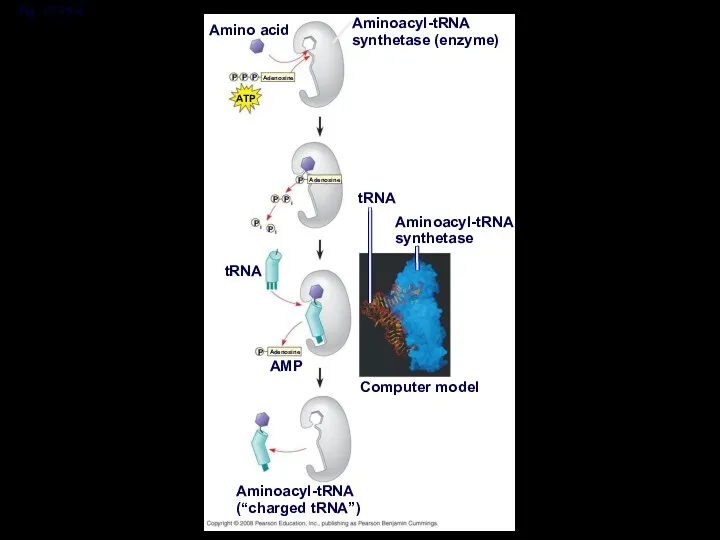
- 34. Fig. 17-15-4 Amino acid Aminoacyl-tRNA synthetase (enzyme) ATP Adenosine P P P Adenosine P P P
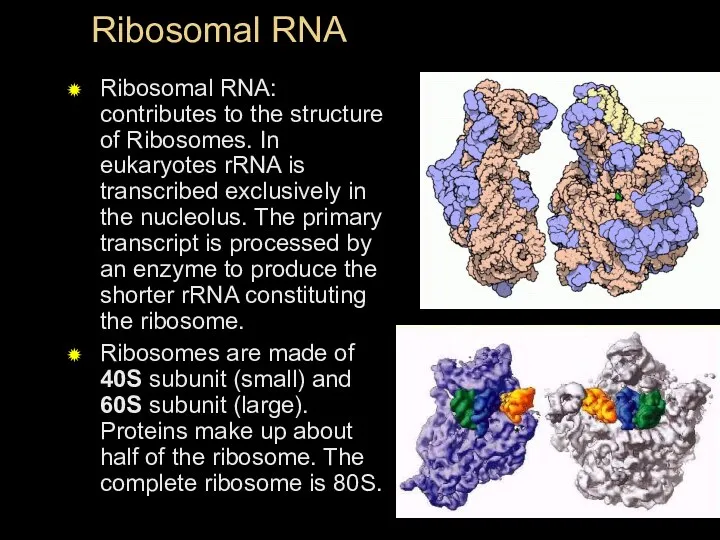
- 37. Ribosomal RNA Ribosomal RNA: contributes to the structure of Ribosomes. In eukaryotes rRNA is transcribed exclusively
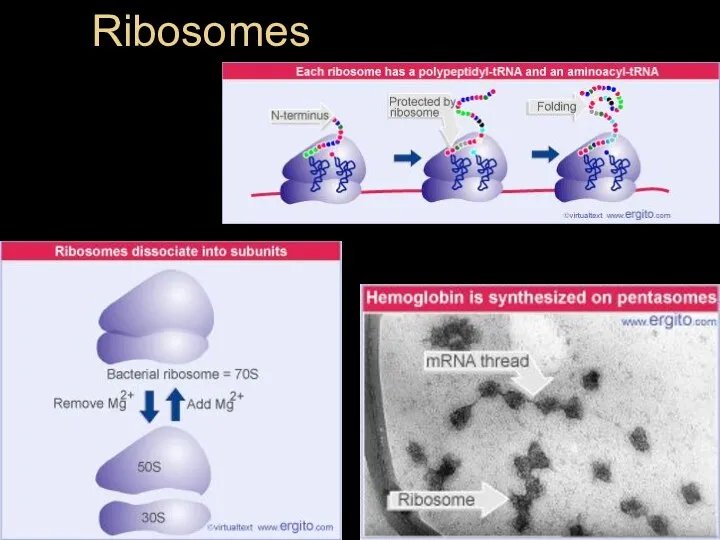
- 38. Ribosomes
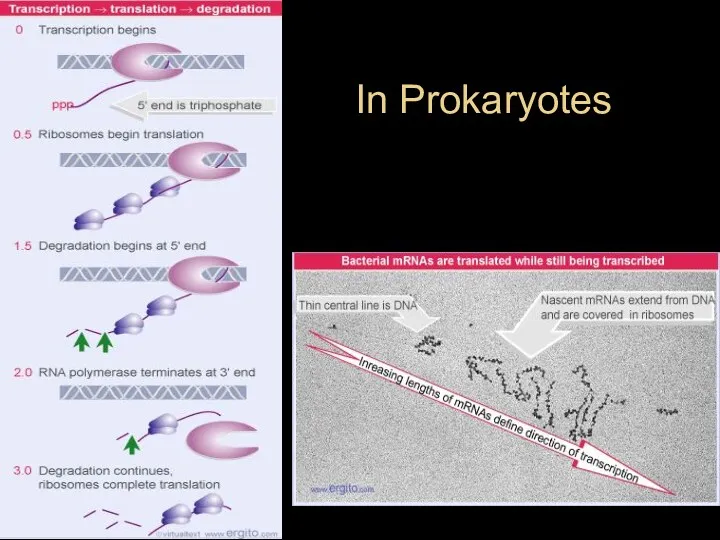
- 40. In Prokaryotes
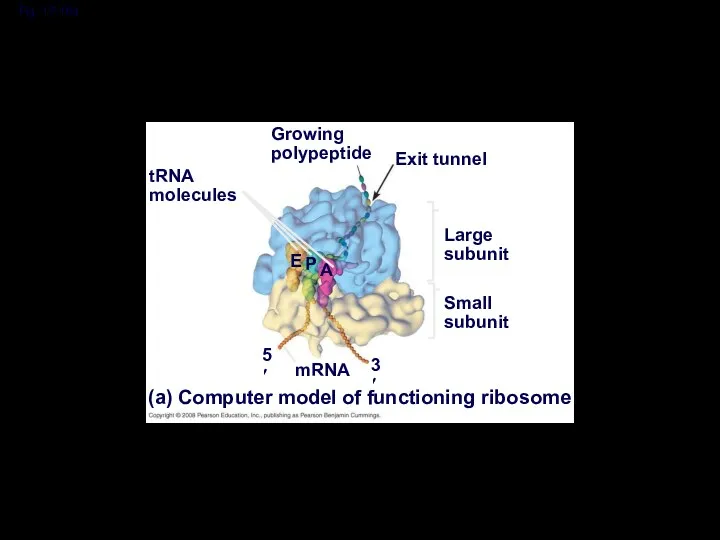
- 41. Fig. 17-16a Growing polypeptide Exit tunnel tRNA molecules Large subunit Small subunit (a) Computer model of
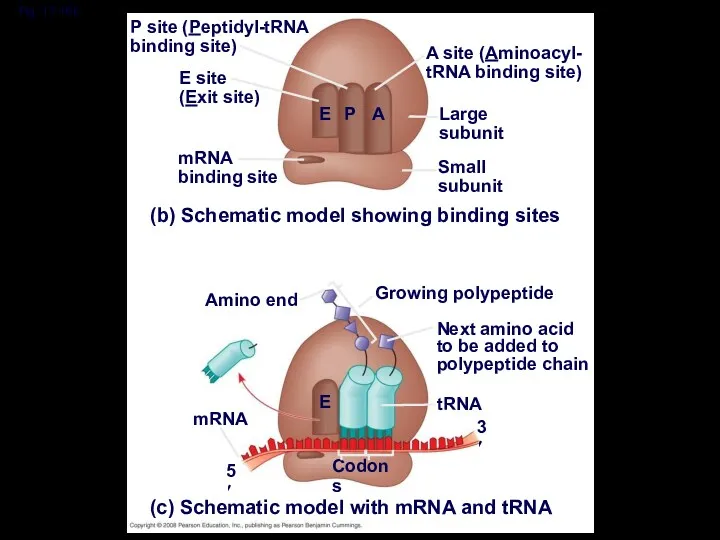
- 42. Fig. 17-16b P site (Peptidyl-tRNA binding site) A site (Aminoacyl- tRNA binding site) E site (Exit
- 43. A ribosome has three binding sites for tRNA: The P site holds the tRNA that carries
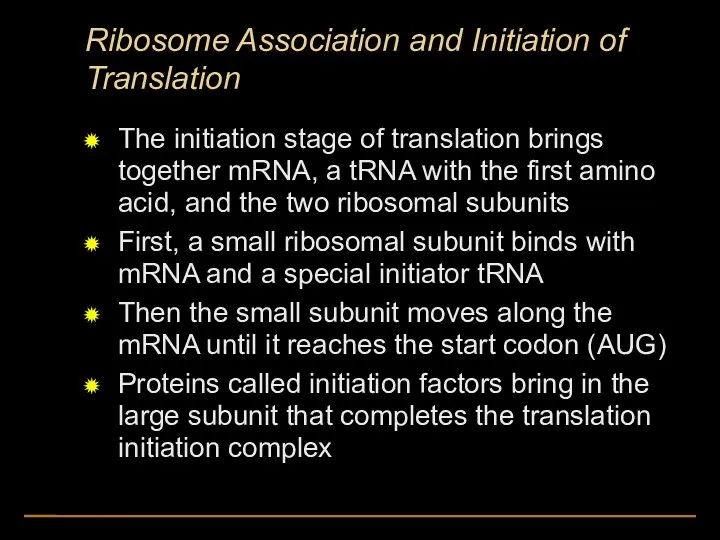
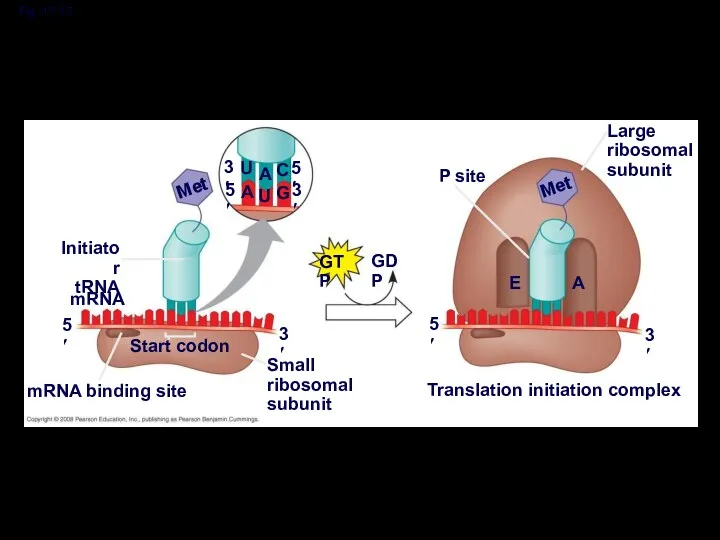
- 44. Ribosome Association and Initiation of Translation The initiation stage of translation brings together mRNA, a tRNA
- 45. Fig. 17-17 3′ 3′ 5′ 5′ U U A A C G Met GTP GDP Initiator
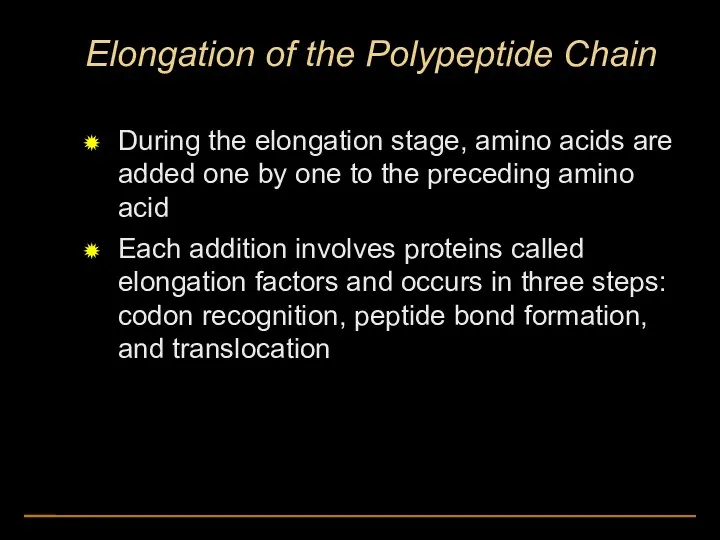
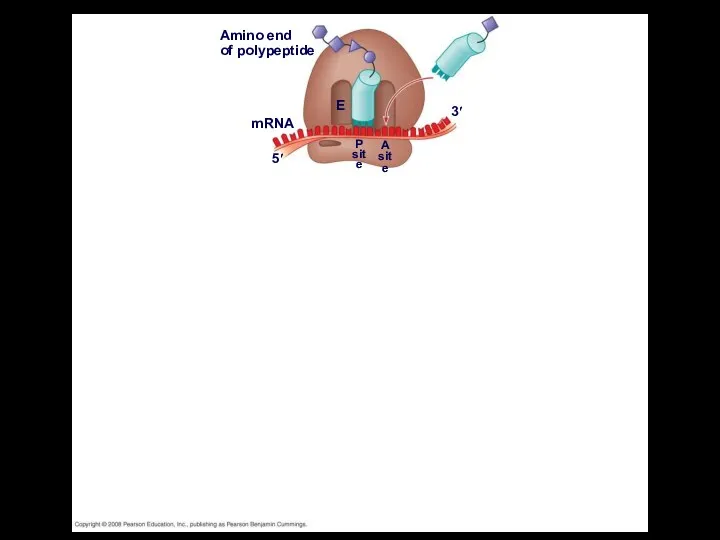
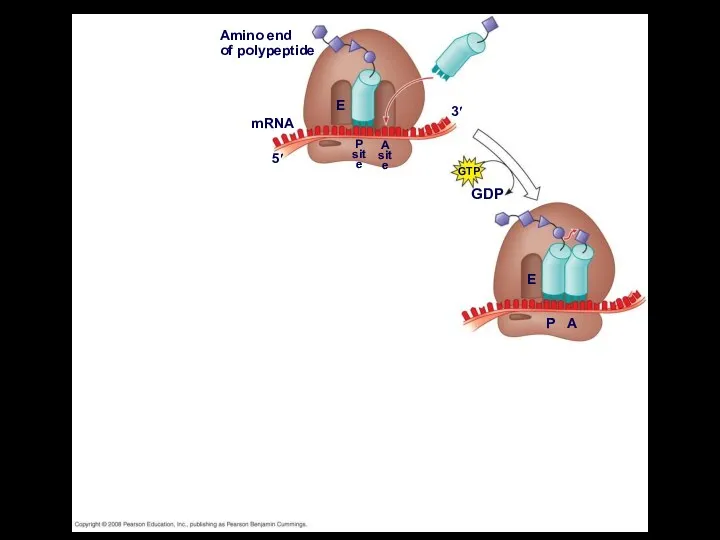
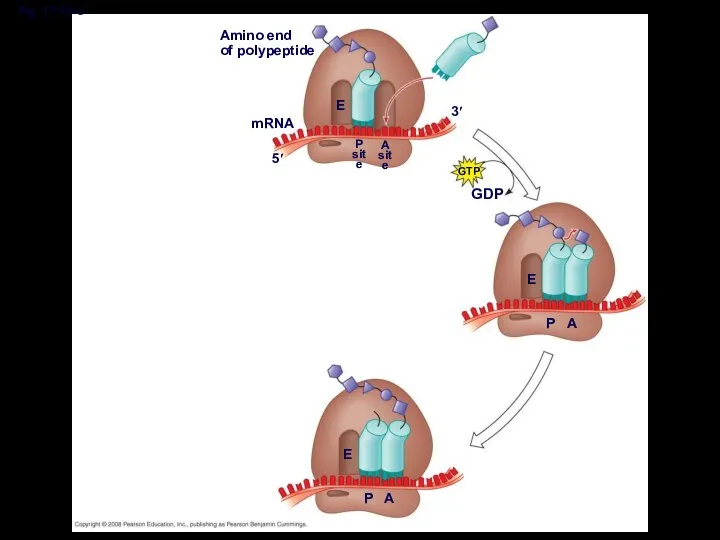
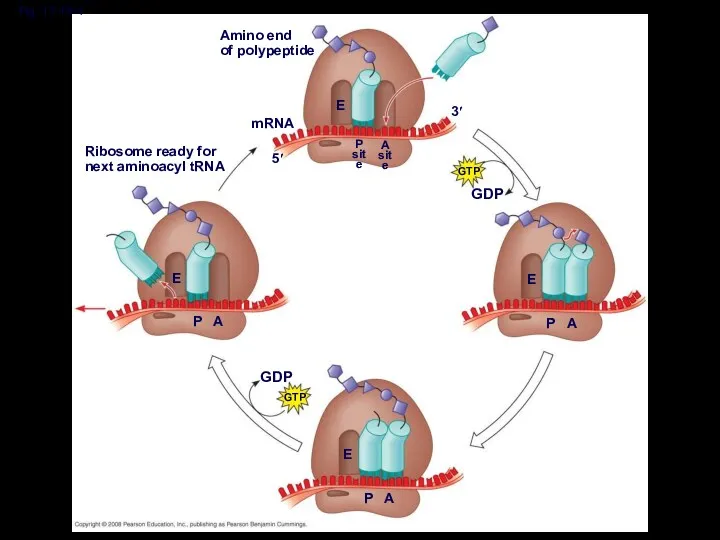
- 46. Elongation of the Polypeptide Chain During the elongation stage, amino acids are added one by one
- 47. Amino end of polypeptide mRNA 5′ 3′ E P site A site
- 48. Amino end of polypeptide mRNA 5′ 3′ E P site A site GTP GDP E P
- 49. Fig. 17-18-3 Amino end of polypeptide mRNA 5′ 3′ E P site A site GTP GDP
- 50. Fig. 17-18-4 Amino end of polypeptide mRNA 5′ 3′ E P site A site GTP GDP
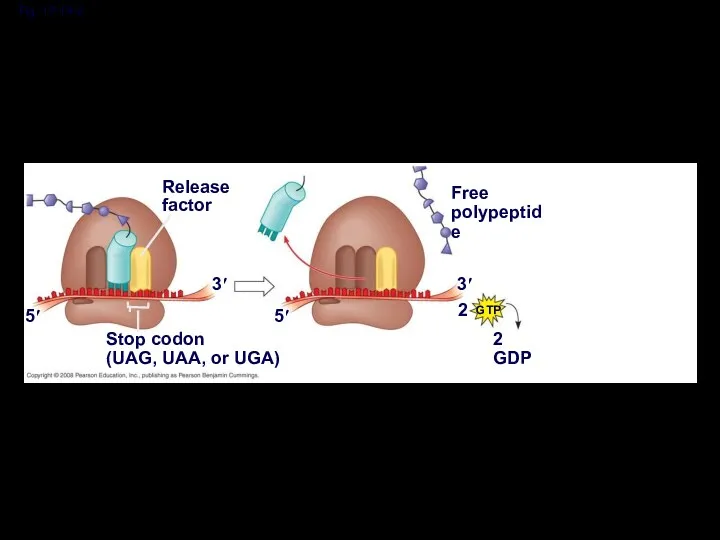
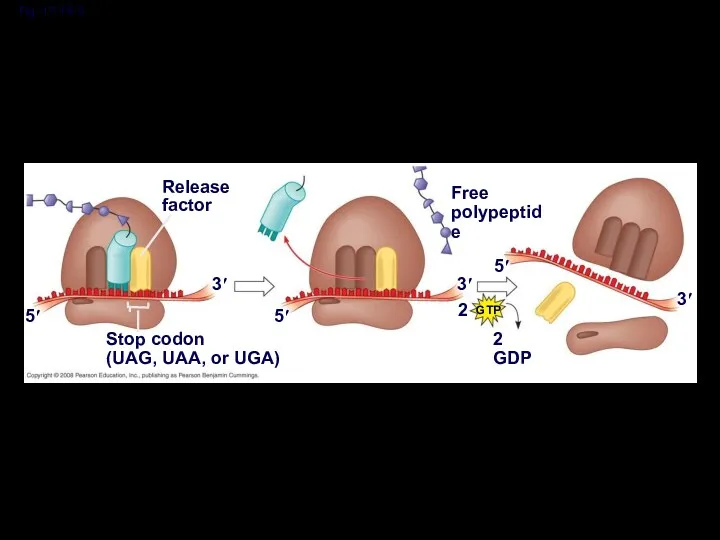
- 51. Termination of Translation Termination occurs when a stop codon in the mRNA reaches the A site
- 52. Fig. 17-19-1 Release factor 3′ 5′ Stop codon (UAG, UAA, or UGA)
- 53. Fig. 17-19-2 Release factor 3′ 5′ Stop codon (UAG, UAA, or UGA) 5′ 3′ 2 Free
- 54. Fig. 17-19-3 Release factor 3′ 5′ Stop codon (UAG, UAA, or UGA) 5′ 3′ 2 Free
- 55. Completing and Targeting the Functional Protein Often translation is not sufficient to make a functional protein
- 56. Protein Folding and Post-Translational Modifications During and after synthesis, a polypeptide chain spontaneously coils and folds
- 57. Targeting Polypeptides to Specific Locations Two populations of ribosomes are evident in cells: free ribsomes (in
- 58. Polypeptide synthesis always begins in the cytosol Synthesis finishes in the cytosol unless the polypeptide signals
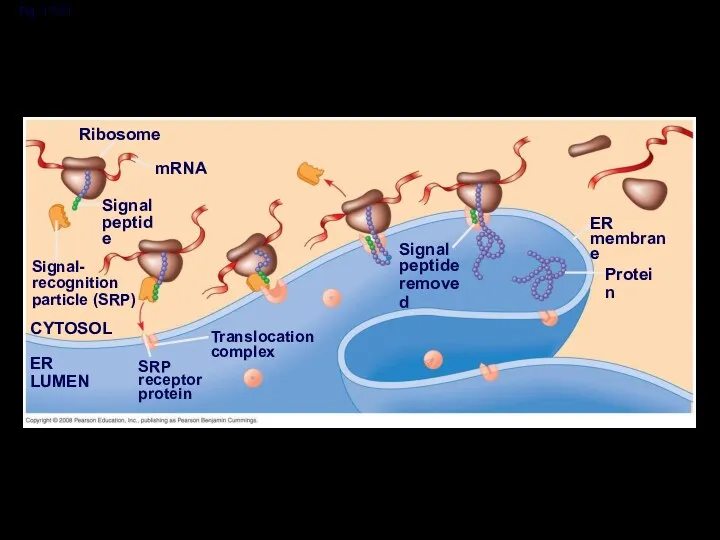
- 59. Fig. 17-21 Ribosome mRNA Signal peptide Signal- recognition particle (SRP) CYTOSOL Translocation complex SRP receptor protein
- 61. Скачать презентацию















 Чудесное превращение. Появление комара
Чудесное превращение. Появление комара Цели, задачи и методы науки селекции
Цели, задачи и методы науки селекции Север - богатый край.
Север - богатый край. Акселерация және ретардация реактивтілік және организмнің резистенттілігі
Акселерация және ретардация реактивтілік және организмнің резистенттілігі Рыбы. Внутреннее строение
Рыбы. Внутреннее строение Загальна характеристика біосфери. Вчення Вернадського про біосферу. Роль живих організмів у біосфері. Біомаса
Загальна характеристика біосфери. Вчення Вернадського про біосферу. Роль живих організмів у біосфері. Біомаса Презентация Дигибридное скрещивание. Третий закон Менделя.
Презентация Дигибридное скрещивание. Третий закон Менделя. Кожа и её производные
Кожа и её производные Отдел Моховидные. Общая характеристика, значение
Отдел Моховидные. Общая характеристика, значение Презентация Значение калия в организме
Презентация Значение калия в организме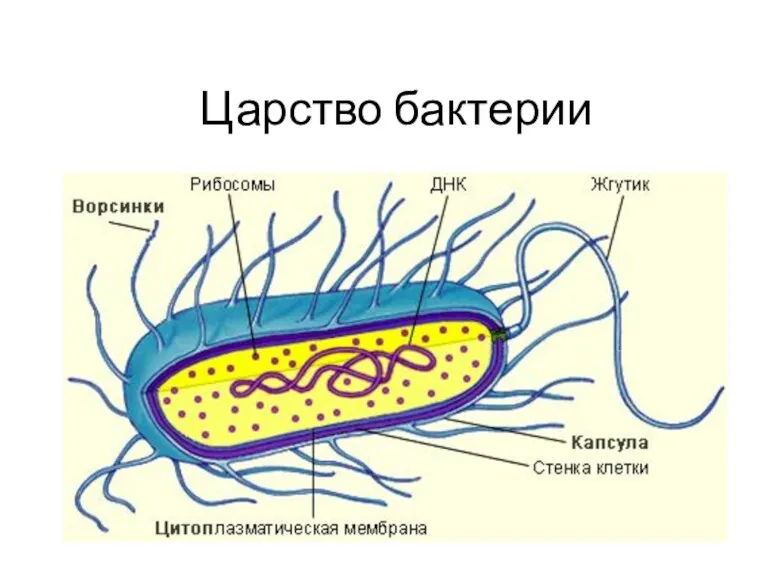 Царство Бактерии. 6 класс
Царство Бактерии. 6 класс презентация Цветок
презентация Цветок Пищевая микробиология. Превращения азотсодержащих веществ
Пищевая микробиология. Превращения азотсодержащих веществ Экология и природопользование. Экосистемы
Экология и природопользование. Экосистемы Кожа. Наружный покров
Кожа. Наружный покров ВИДЫ ВЕГЕТАТИВНОГО РАЗМНОЖЕНИЯ РАСТЕНИИ
ВИДЫ ВЕГЕТАТИВНОГО РАЗМНОЖЕНИЯ РАСТЕНИИ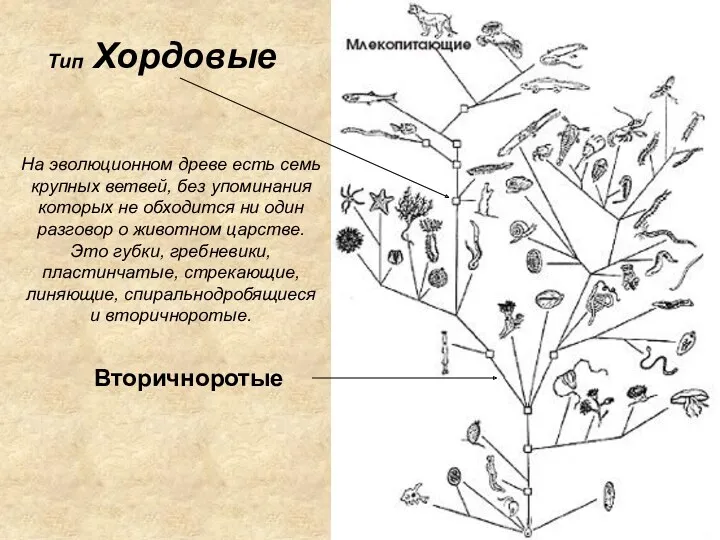 Тип Хордовые
Тип Хордовые Какие бывают животные? Окружающий мир. 2 класс
Какие бывают животные? Окружающий мир. 2 класс Вегетативное размножение растений
Вегетативное размножение растений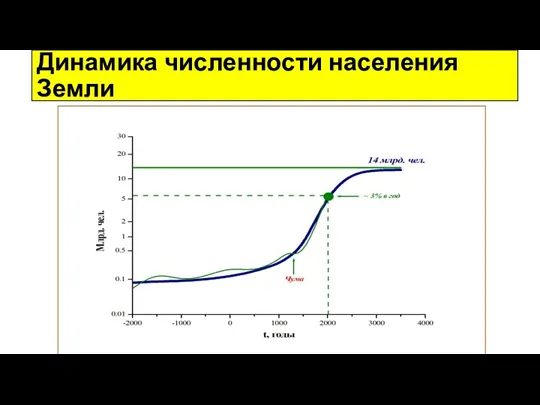 Динамика численности населения Земли
Динамика численности населения Земли Осторожно, борщевик!
Осторожно, борщевик! Среды обитания организмов
Среды обитания организмов Малина звичайна
Малина звичайна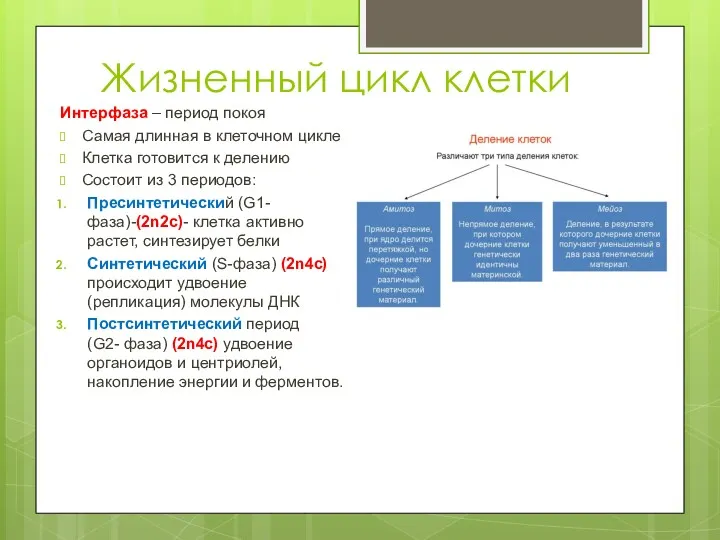 Жизненный цикл клетки
Жизненный цикл клетки Болотные птицы
Болотные птицы Общая характеристика грибов. Многообразие
Общая характеристика грибов. Многообразие Растения болот
Растения болот Международная классификация микроорганизмов по Берги
Международная классификация микроорганизмов по Берги