Содержание
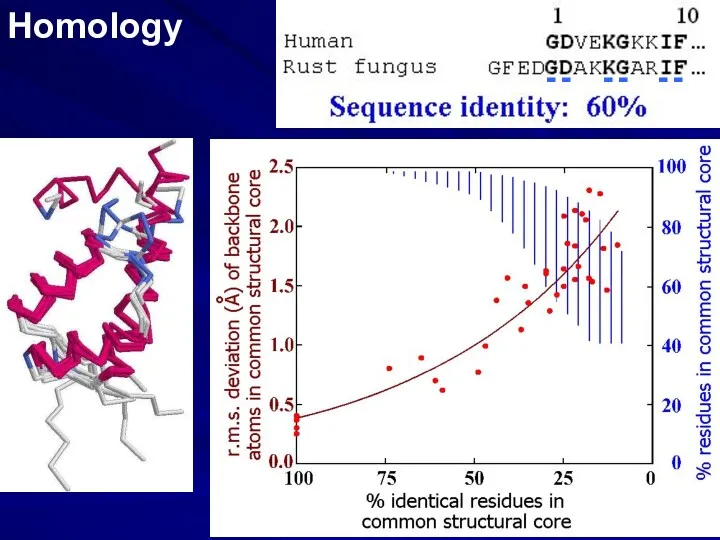
- 2. Homology - - - - - -
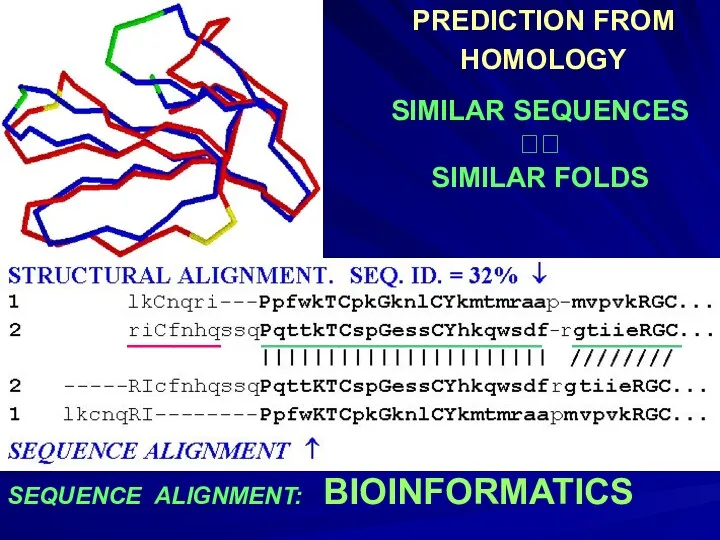
- 3. SEQUENCE ALIGNMENT: BIOINFORMATICS PREDICTION FROM HOMOLOGY SIMILAR SEQUENCES ?? SIMILAR FOLDS ______ __________________ _______
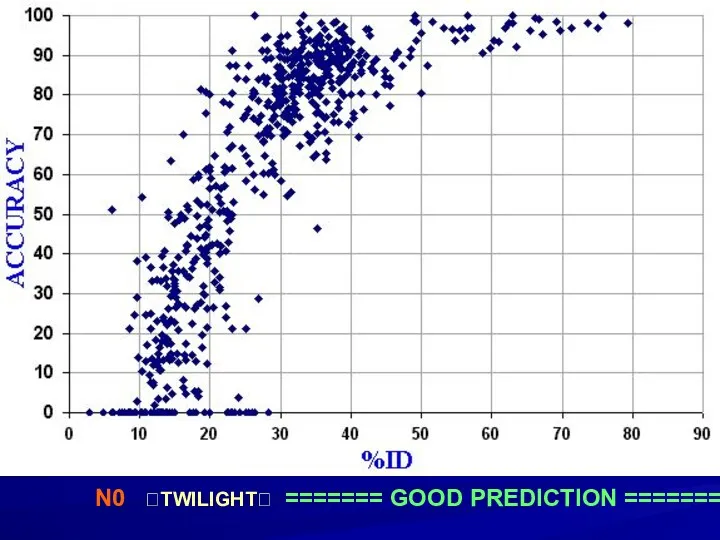
- 4. N0 ?TWILIGHT? ======= GOOD PREDICTION =======
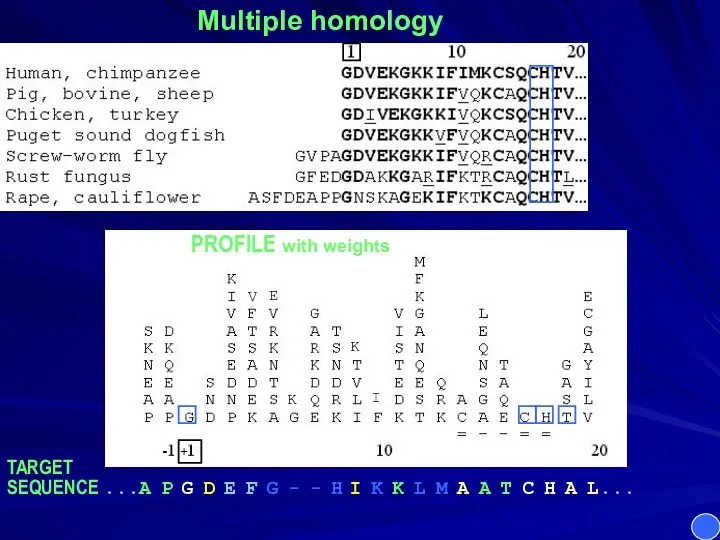
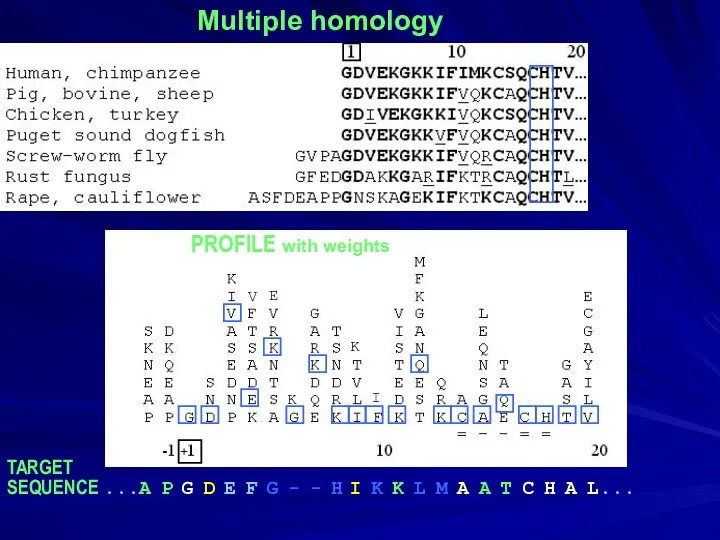
- 5. Multiple homology PROFILE with weights TARGET SEQUENCE ...A P G D E F G - -
- 6. Multiple homology PROFILE with weights TARGET SEQUENCE ...A P G D E F G - -
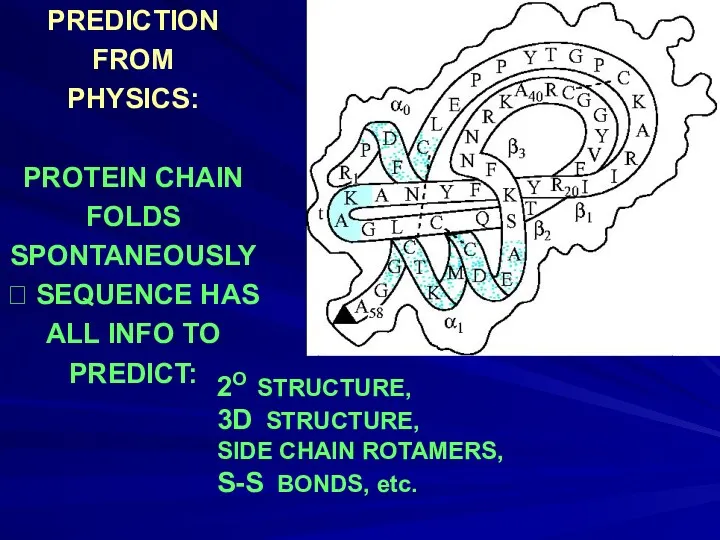
- 7. PREDICTION FROM PHYSICS: PROTEIN CHAIN FOLDS SPONTANEOUSLY ? SEQUENCE HAS ALL INFO TO PREDICT: 2O STRUCTURE,
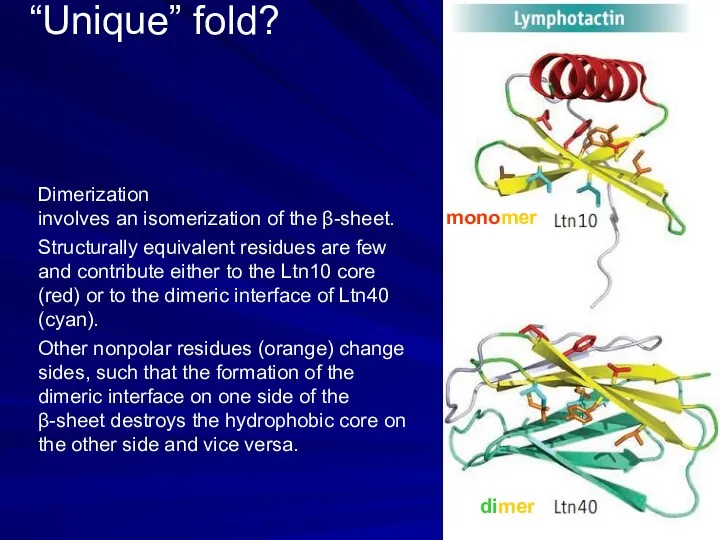
- 8. “Unique” fold? monomer dimer Dimerization involves an isomerization of the β-sheet. Structurally equivalent residues are few
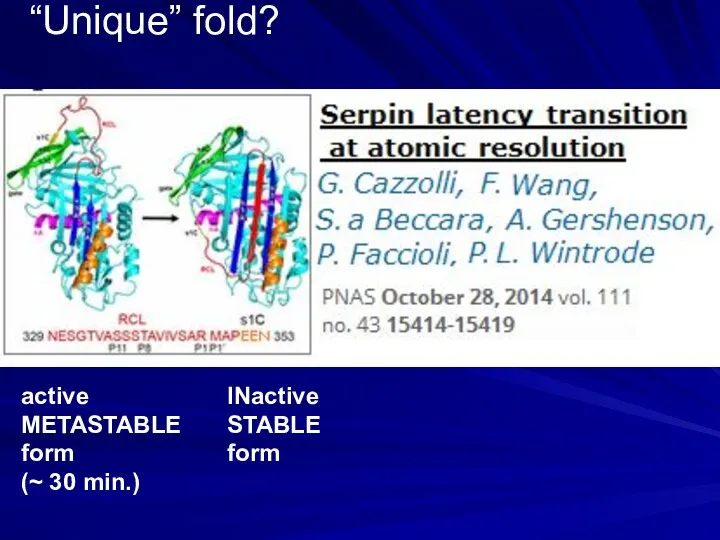
- 9. “Unique” fold? active METASTABLE form (~ 30 min.) INactive STABLE form
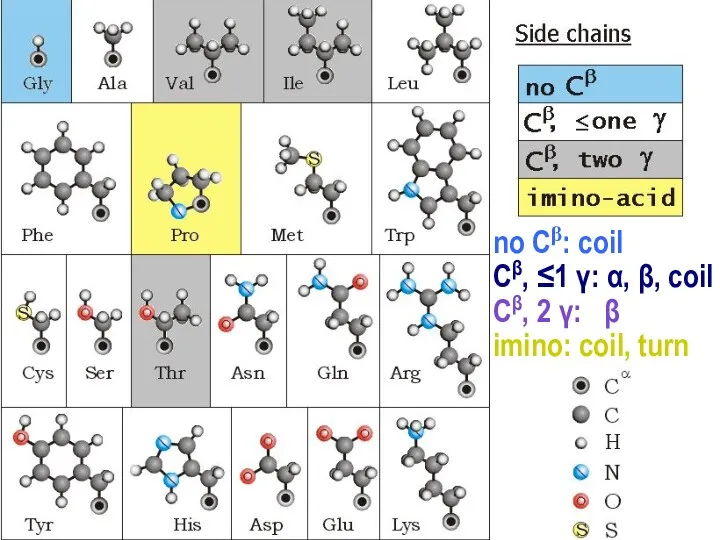
- 10. no Cβ: coil Сβ, ≤1 γ: α, β, coil Сβ, 2 γ: β imino: coil, turn
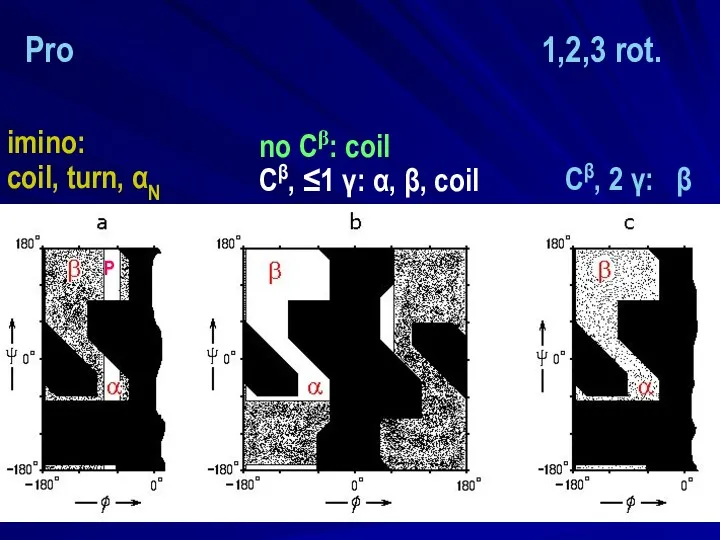
- 11. no Сβ: coil Сβ, ≤1 γ: α, β, coil imino: coil, turn, αN Сβ, 2 γ:
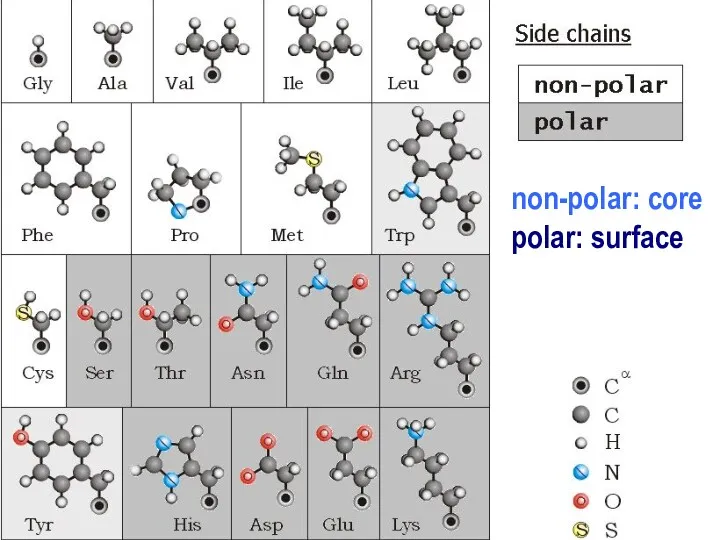
- 12. non-polar: core polar: surface
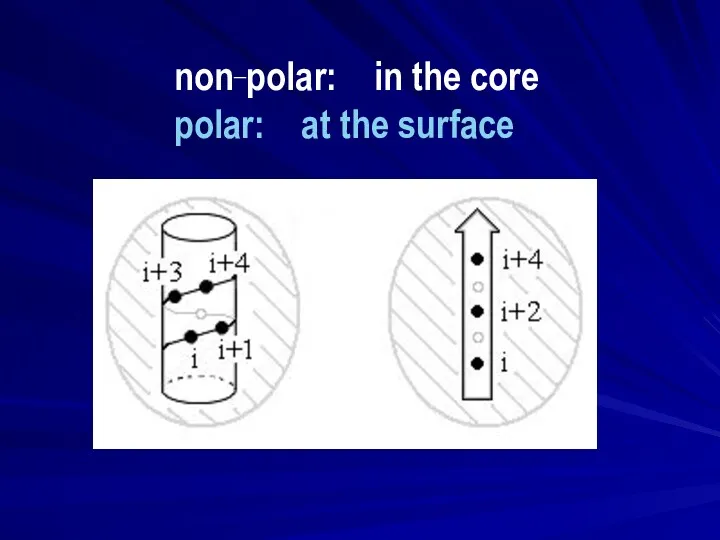
- 13. non_polar: in the core polar: at the surface
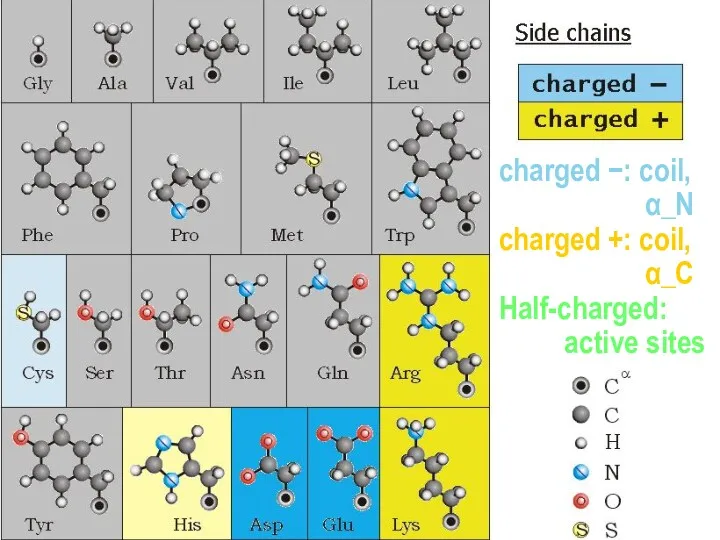
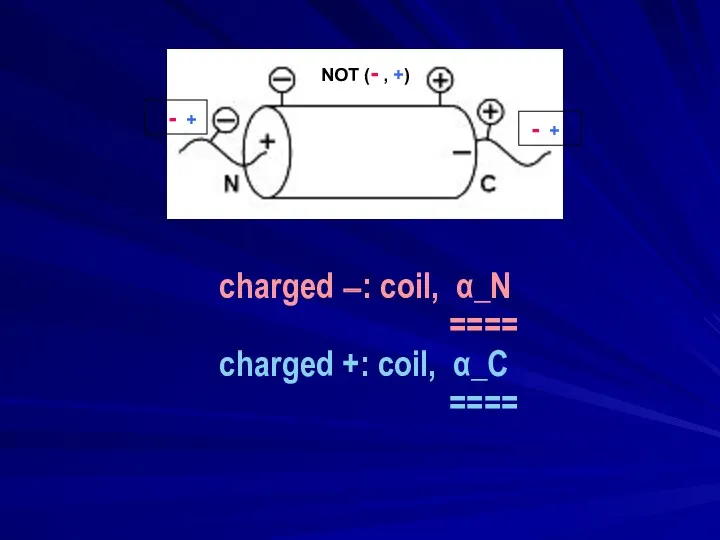
- 14. charged −: coil, α_N charged +: coil, α_C Half-charged: active sites
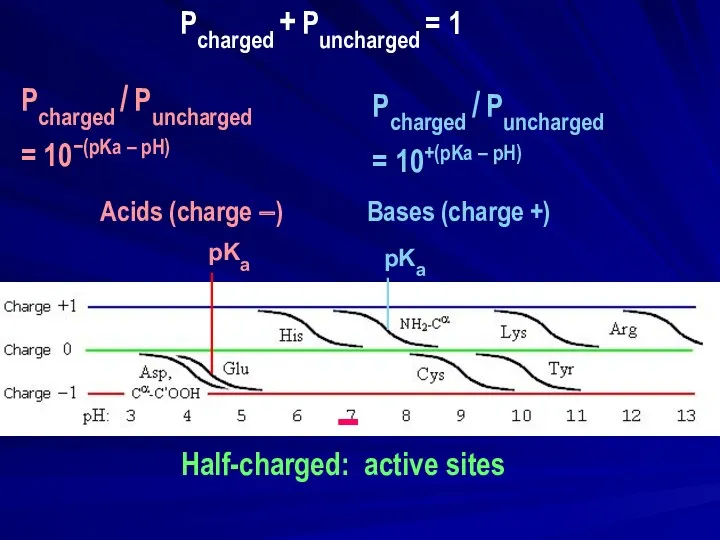
- 15. − Half-charged: active sites pKa | | pKa | | | | Pcharged / Puncharged =
- 16. charged −: coil, α_N ==== charged +: coil, α_C ==== - + - + NOT (-
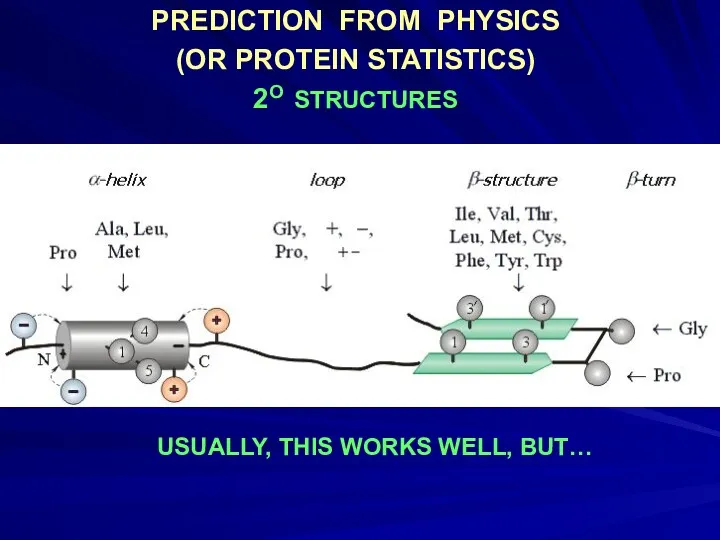
- 17. PREDICTION FROM PHYSICS (OR PROTEIN STATISTICS) 2O STRUCTURES USUALLY, THIS WORKS WELL, BUT…
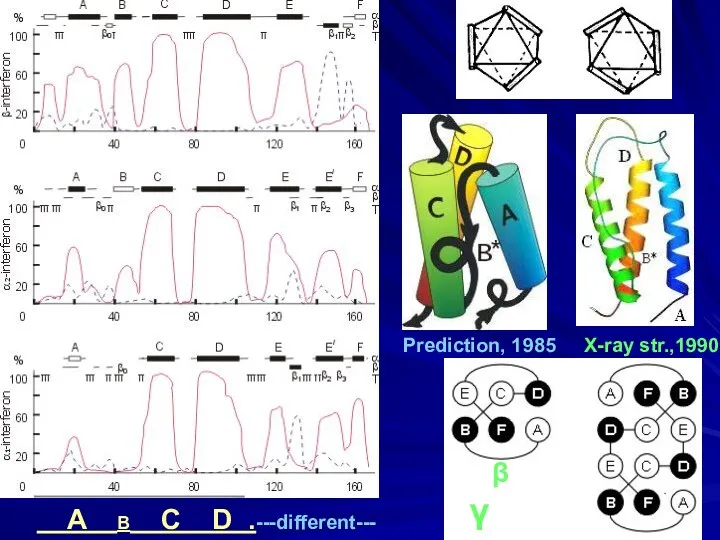
- 18. A B C D .---different--- Prediction, 1985 X-ray str.,1990 β γ
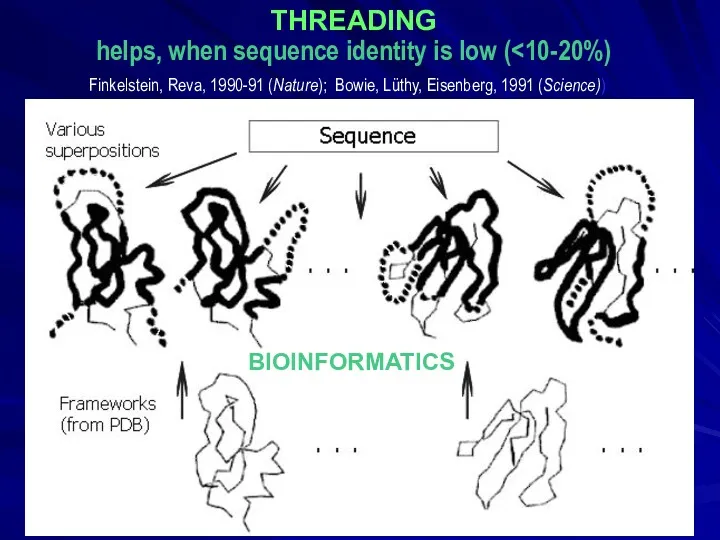
- 19. THREADING helps, when sequence identity is low ( BIOINFORMATICS Finkelstein, Reva, 1990-91 (Nature); Bowie, Lüthy, Eisenberg,
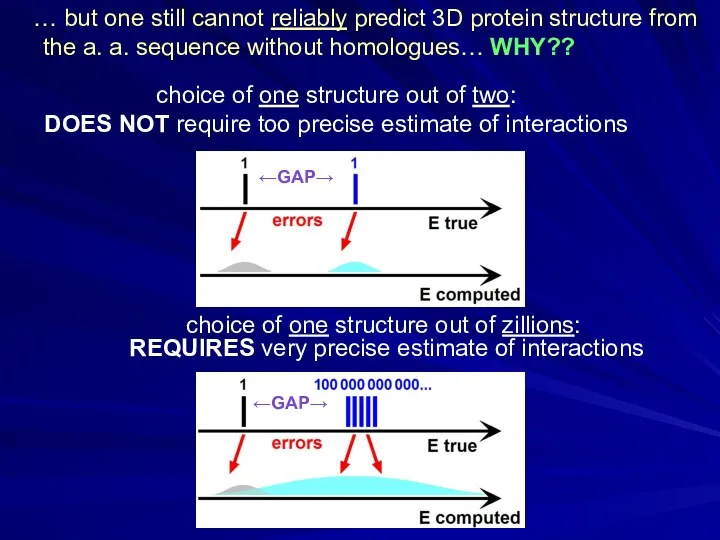
- 20. choice of one structure out of zillions: REQUIRES very precise estimate of interactions choice of one
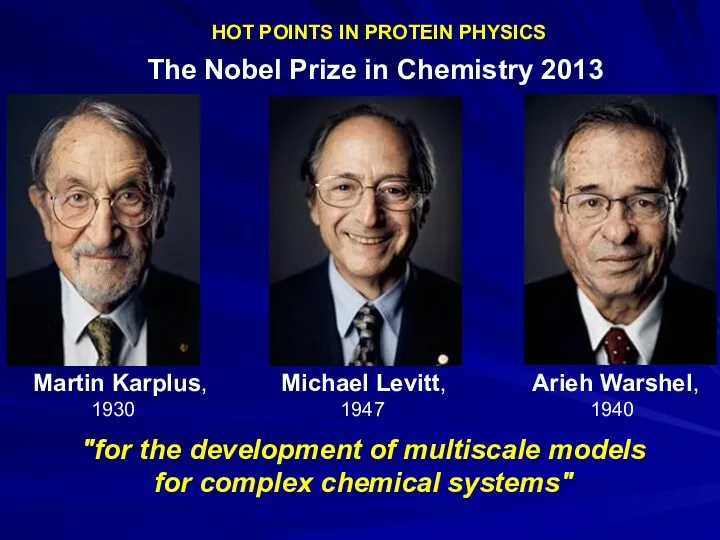
- 21. HOT POINTS IN PROTEIN PHYSICS The Nobel Prize in Chemistry 2013 Martin Karplus, Michael Levitt, Arieh
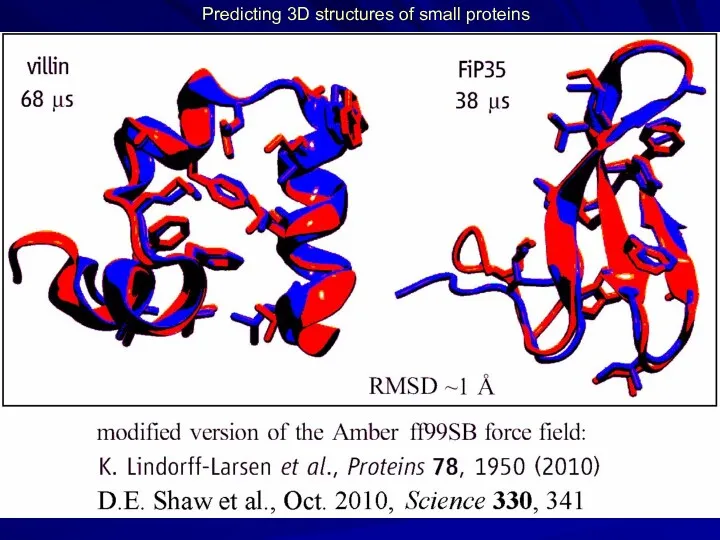
- 22. Predicting 3D structures of small proteins
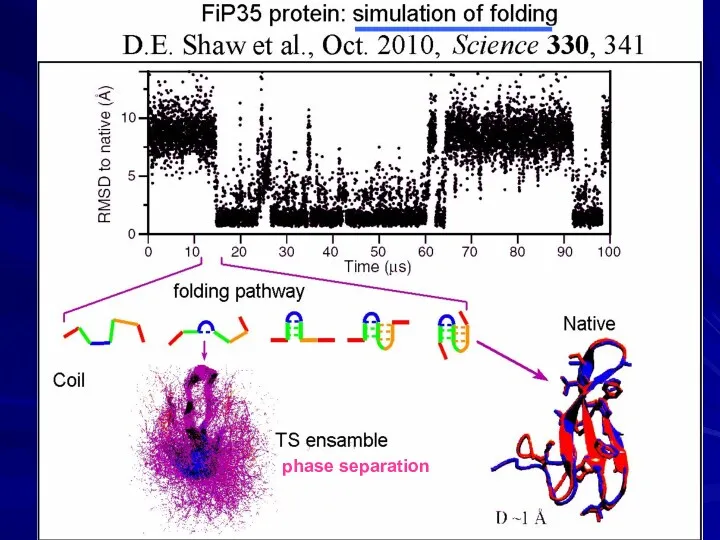
- 23. HOT POINTS IN PROTEIN PHYSICS David E. Shaw, 1951 “D. E. Shaw Research” US$ 3.5 billion
- 24. phase separation
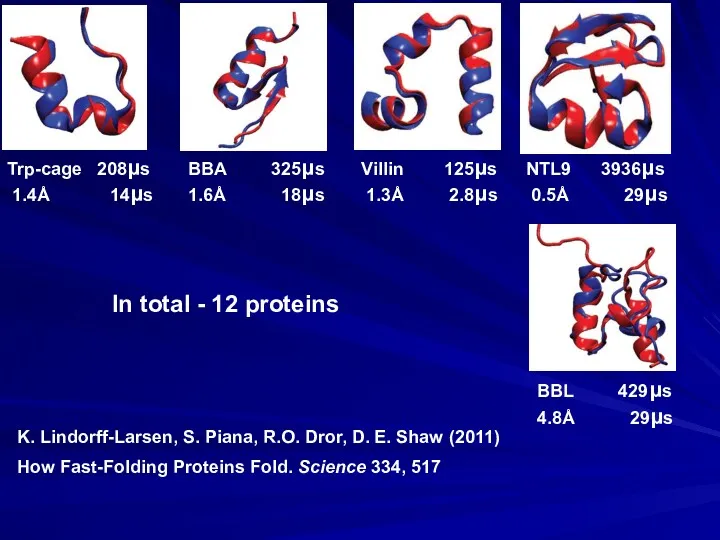
- 25. How Fast-Folding Proteins Fold. Science 334, 517 K. Lindorff-Larsen, S. Piana, R.O. Dror, D. E. Shaw
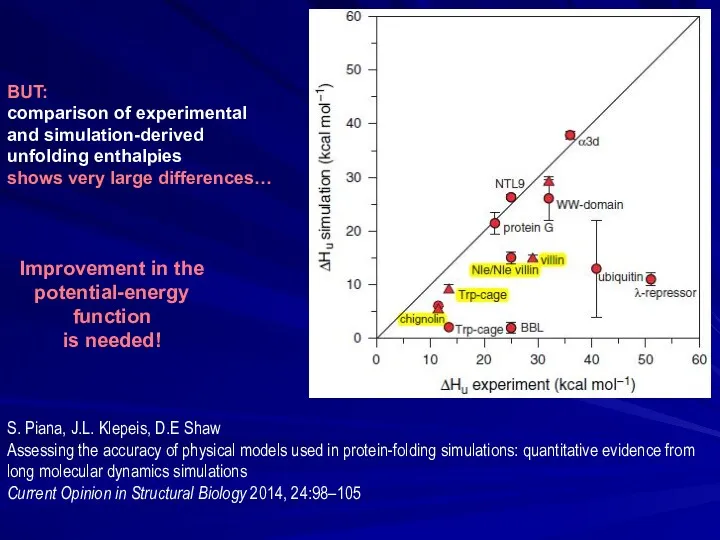
- 26. BUT: comparison of experimental and simulation-derived unfolding enthalpies shows very large differences… S. Piana, J.L. Klepeis,
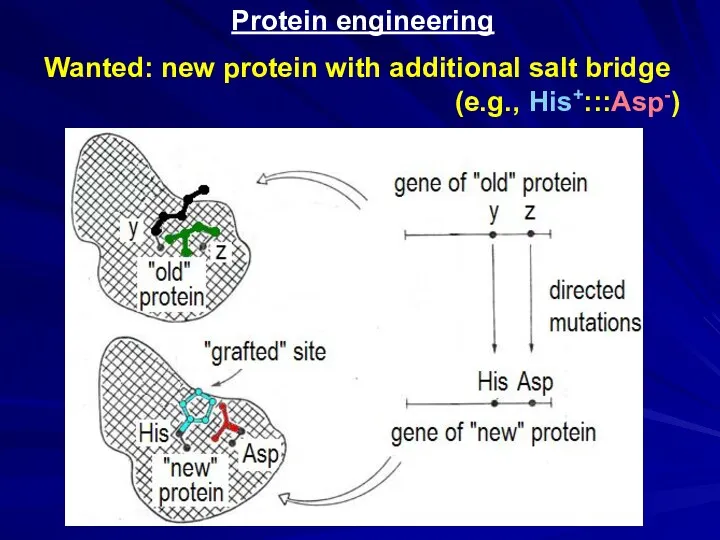
- 27. Protein engineering Wanted: new protein with additional salt bridge (e.g., His+:::Asp-)
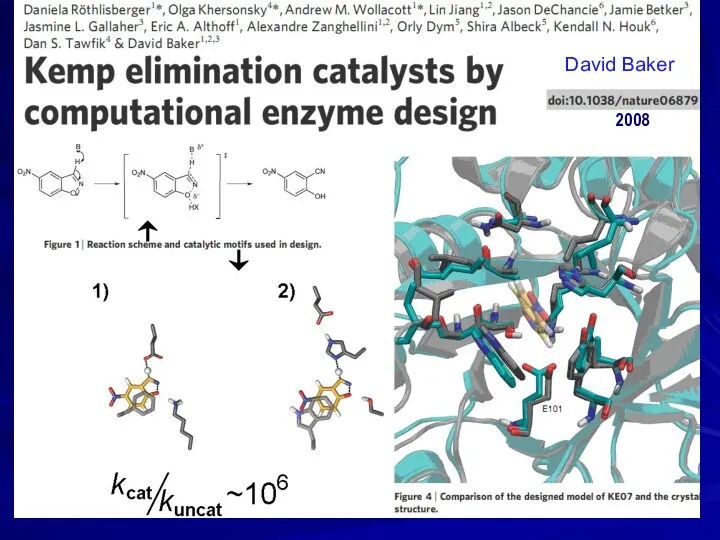
- 28. 2008 David Baker
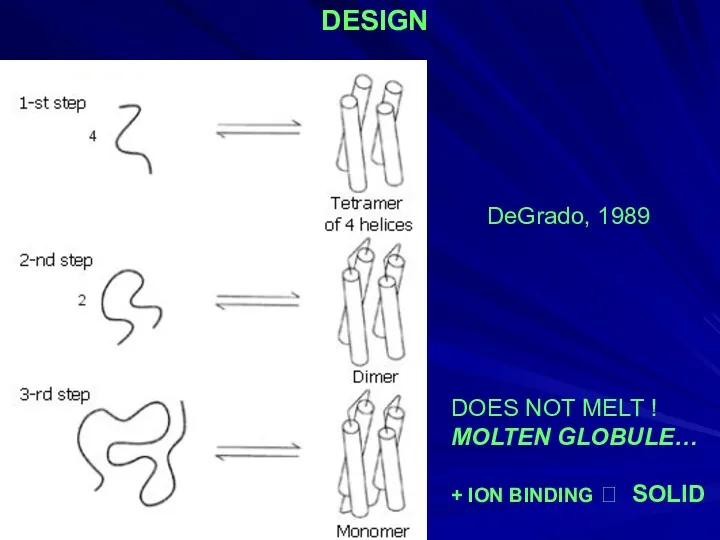
- 29. DOES NOT MELT ! MOLTEN GLOBULE… + ION BINDING ? SOLID DeGrado, 1989 DESIGN
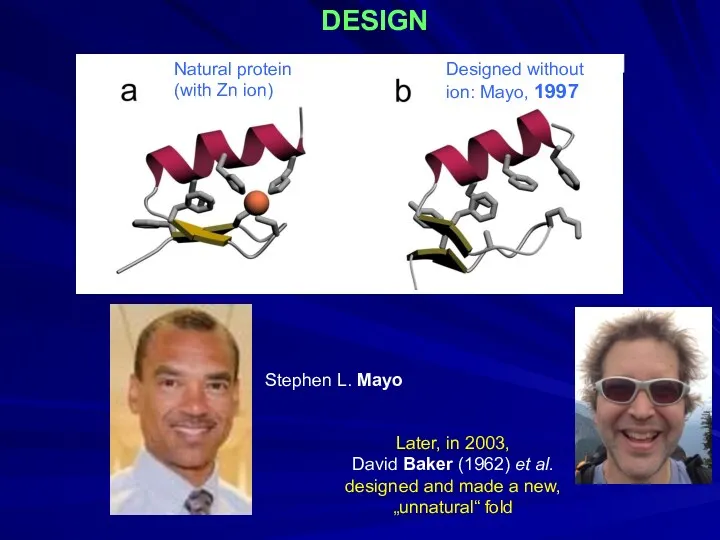
- 30. DESIGN Designed without ion: Mayo, 1997 Natural protein (with Zn ion) Stephen L. Mayo Later, in
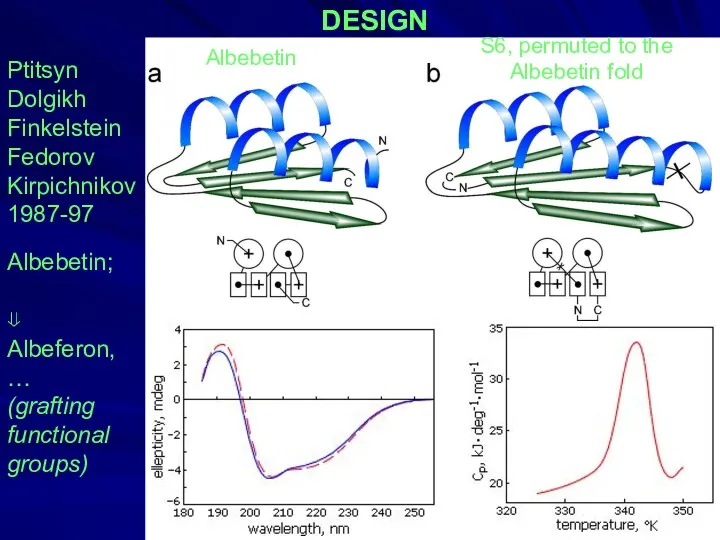
- 31. DESIGN Ptitsyn Dolgikh Finkelstein Fedorov Kirpichnikov 1987-97 Albebetin; ⇓ Albeferon, … (grafting functional groups) Albebetin S6,
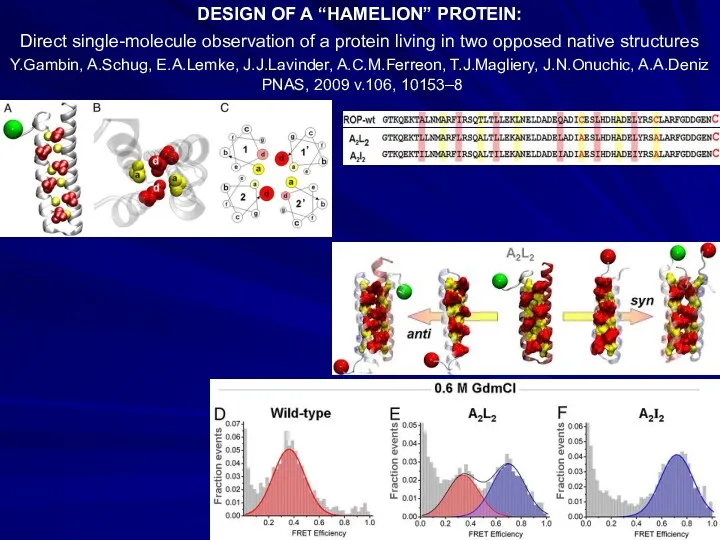
- 32. DESIGN OF A “HAMELION” PROTEIN: Direct single-molecule observation of a protein living in two opposed native
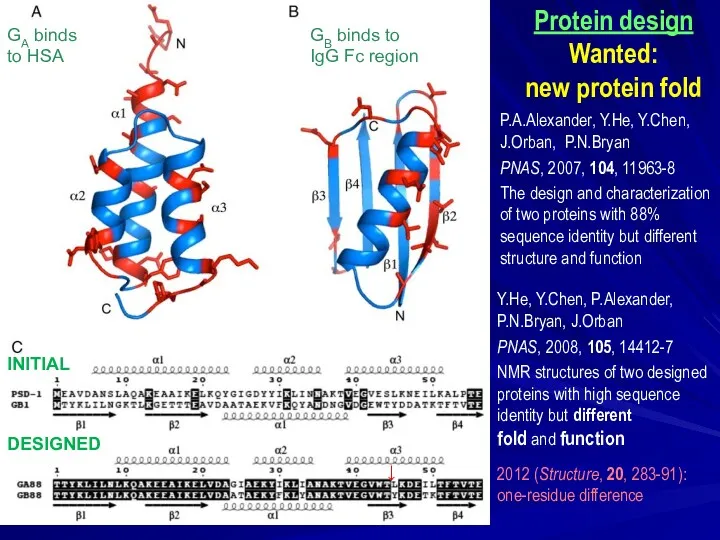
- 33. Y.He, Y.Chen, P.Alexander, P.N.Bryan, J.Orban PNAS, 2008, 105, 14412-7 NMR structures of two designed proteins with
- 35. Скачать презентацию

 Электромагниттік сәуле шығару
Электромагниттік сәуле шығару Особенности заданий ЕГЭ. Колебания и волны
Особенности заданий ЕГЭ. Колебания и волны Линзы. Построение изображения в собирающей линзе
Линзы. Построение изображения в собирающей линзе Остов, кривошипно-шатунний та газорозподільний механізми ДВЗ. (Лекція 3.1)
Остов, кривошипно-шатунний та газорозподільний механізми ДВЗ. (Лекція 3.1) Структура и принципы интеграции МС
Структура и принципы интеграции МС Диэлектрики в электрическом поле
Диэлектрики в электрическом поле Перемещение при прямолинейном равномерном движении
Перемещение при прямолинейном равномерном движении Основные положения молекулярно-кинетической теории и их опытные подтверждения
Основные положения молекулярно-кинетической теории и их опытные подтверждения Электронная проводимость металлов. Электрический ток и его характеристики
Электронная проводимость металлов. Электрический ток и его характеристики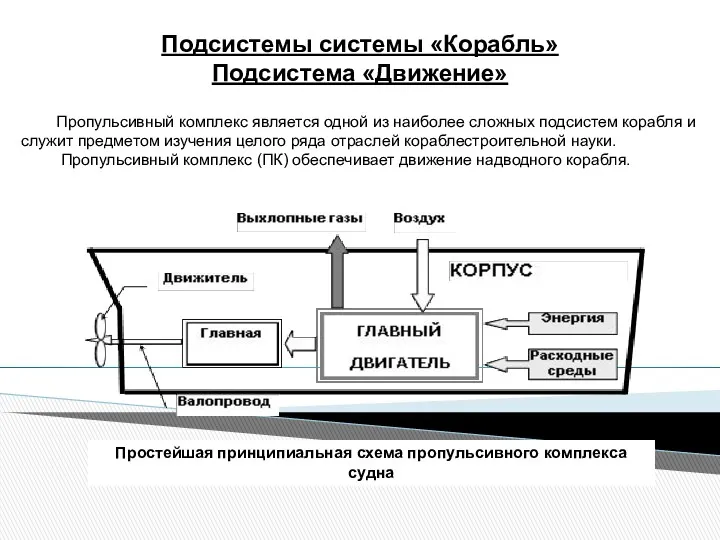 Подсистемы системы Корабль. Подсистема Движение
Подсистемы системы Корабль. Подсистема Движение Известные физики
Известные физики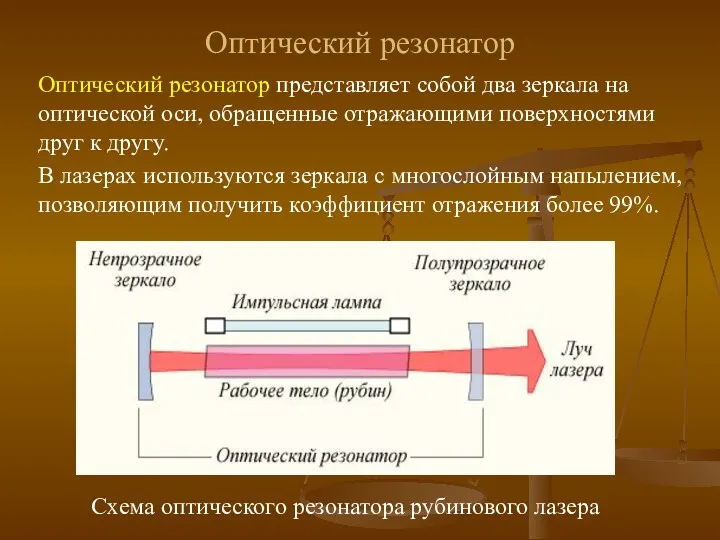 Оптический резонатор
Оптический резонатор Презентация 10 класса Газовые законы
Презентация 10 класса Газовые законы Специальная теория относительности
Специальная теория относительности Презентация Интерференция света
Презентация Интерференция света Презентация доклада системнодеятельностного подхода на уроках физики
Презентация доклада системнодеятельностного подхода на уроках физики Уравнения Максвелла
Уравнения Максвелла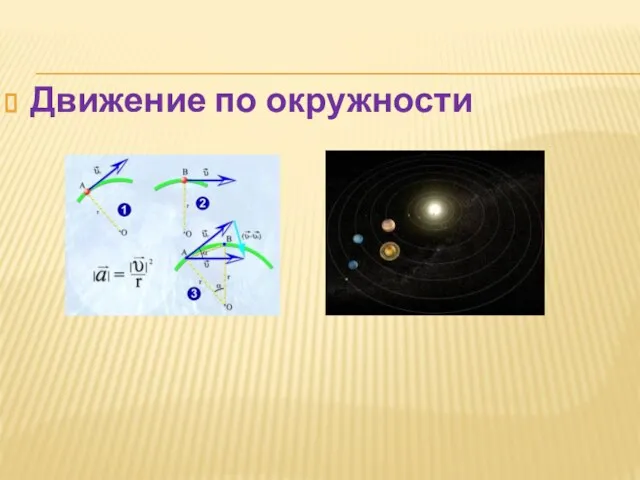 Движение по окружности
Движение по окружности Занятие № 1 Методы повышения эффективности усвоения понятийпри изучении темы:Кинематика
Занятие № 1 Методы повышения эффективности усвоения понятийпри изучении темы:Кинематика Детекторы нейтронов
Детекторы нейтронов Презентация 8 класс. Количество теплоты. Едельная теплоемкость
Презентация 8 класс. Количество теплоты. Едельная теплоемкость Водородная бомба
Водородная бомба Решение физических задач с применением производной функции
Решение физических задач с применением производной функции Презентация Броуновское движение. Диффузия. Взаимодействие молекул
Презентация Броуновское движение. Диффузия. Взаимодействие молекул Сила трения. Особенности сил трения
Сила трения. Особенности сил трения Способы герметизации клепаных швов и изделий
Способы герметизации клепаных швов и изделий Урок и презентация по теме Плавание тел 7 класс
Урок и презентация по теме Плавание тел 7 класс Биологическое действие радиации. Закон радиоактивного распада
Биологическое действие радиации. Закон радиоактивного распада