Содержание
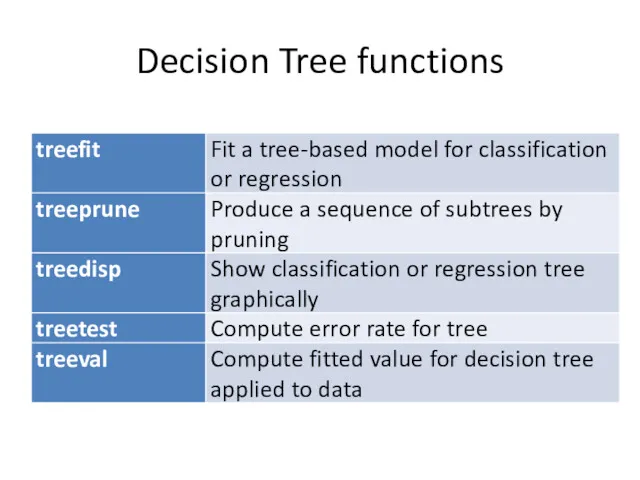
- 2. Decision Tree functions
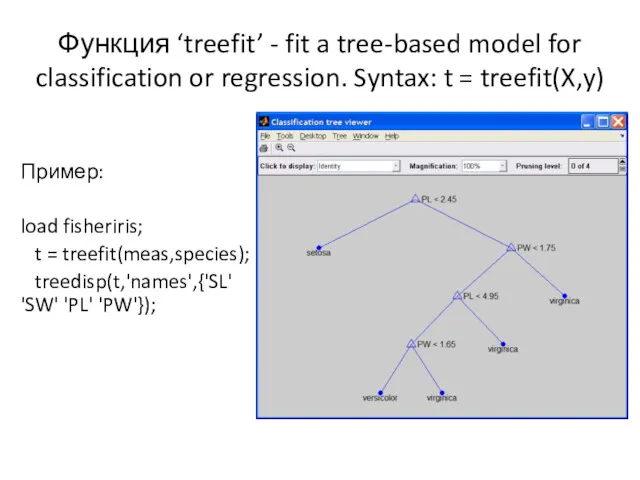
- 3. Функция ‘treefit’ - fit a tree-based model for classification or regression. Syntax: t = treefit(X,y) Пример:
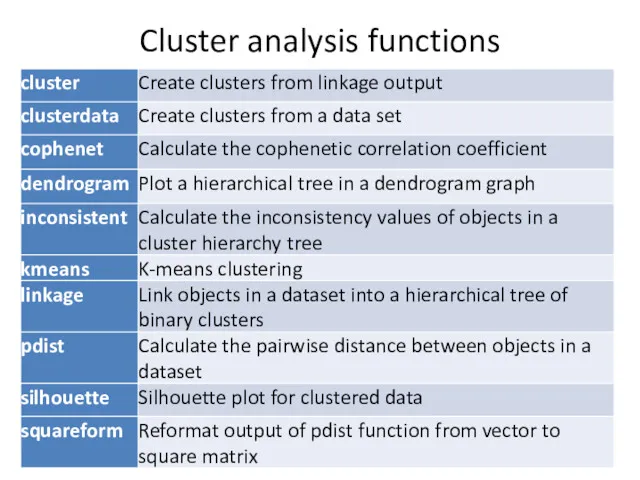
- 4. Cluster analysis functions
- 5. Функция kmeans IDX = kmeans(X,k) [IDX,C] = kmeans(X,k) [IDX,C,sumd] = kmeans(X,k) [IDX,C,sumd,D] = kmeans(X,k) [...] =
- 6. Параметр ‘distance’ 'sqEuclidean‘ - Squared Euclidean distance (default). 'cityblock‘ - Sum of absolute differences, i.e., L1.
- 7. Параметр ‘start’ Method used to choose the initial cluster centroid positions, sometimes known as "seeds". Valid
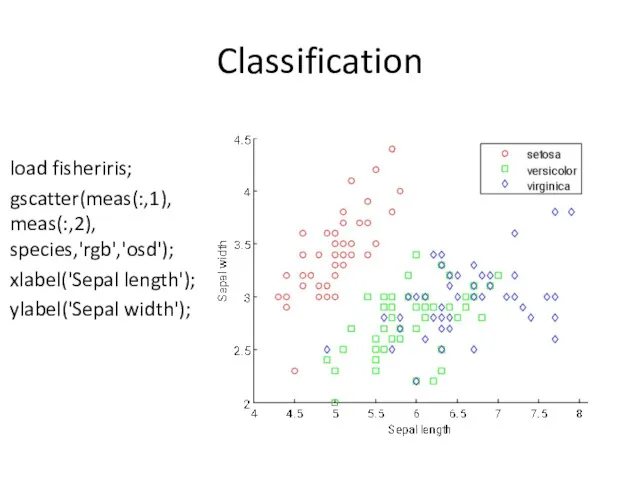
- 8. Classification load fisheriris; gscatter(meas(:,1), meas(:,2), species,'rgb','osd'); xlabel('Sepal length'); ylabel('Sepal width');
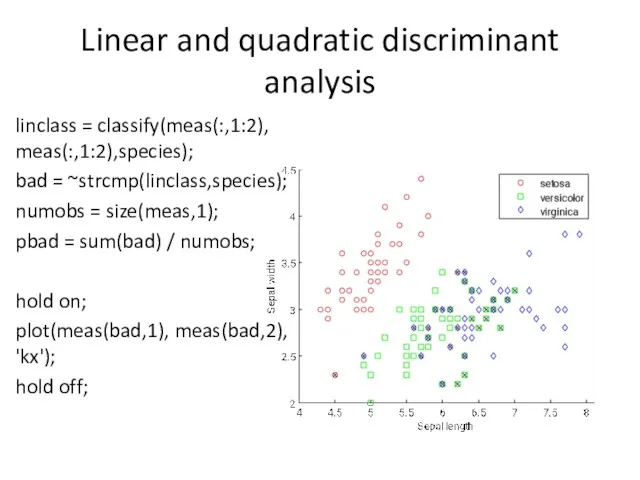
- 9. Linear and quadratic discriminant analysis linclass = classify(meas(:,1:2), meas(:,1:2),species); bad = ~strcmp(linclass,species); numobs = size(meas,1); pbad
- 10. Visualization regioning the plane [x,y] = meshgrid(4:.1:8,2:.1:4.5); x = x(:); y = y(:); j = classify([x
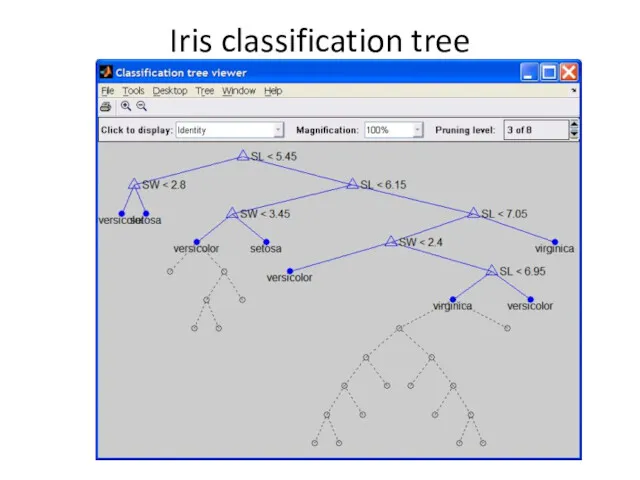
- 11. Decision trees tree = treefit(meas(:,1:2), species); [dtnum,dtnode,dtclass] = treeval(tree, meas(:,1:2)); bad = ~strcmp(dtclass,species); sum(bad) / numobs
- 12. Iris classification tree
- 13. Тестирование качества классификации resubcost = treetest(tree,'resub'); [cost,secost,ntermnodes,bestlevel] = treetest(tree,'cross',meas(:,1:2),species); plot(ntermnodes,cost,'b-', ntermnodes,resubcost,'r--') figure(gcf); xlabel('Number of terminal nodes');
- 14. Выбор уровня [mincost,minloc] = min(cost); cutoff = mincost + secost(minloc); hold on plot([0 20], [cutoff cutoff],
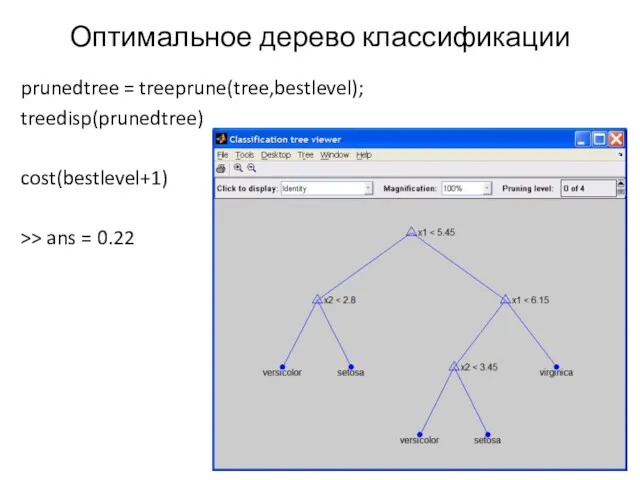
- 15. Оптимальное дерево классификации prunedtree = treeprune(tree,bestlevel); treedisp(prunedtree) cost(bestlevel+1) >> ans = 0.22
- 17. Скачать презентацию
![Функция kmeans IDX = kmeans(X,k) [IDX,C] = kmeans(X,k) [IDX,C,sumd] =](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/150355/slide-4.jpg)


![Visualization regioning the plane [x,y] = meshgrid(4:.1:8,2:.1:4.5); x = x(:);](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/150355/slide-9.jpg)
![Decision trees tree = treefit(meas(:,1:2), species); [dtnum,dtnode,dtclass] = treeval(tree, meas(:,1:2)); bad = ~strcmp(dtclass,species); sum(bad) / numobs](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/150355/slide-10.jpg)
![Тестирование качества классификации resubcost = treetest(tree,'resub'); [cost,secost,ntermnodes,bestlevel] = treetest(tree,'cross',meas(:,1:2),species); plot(ntermnodes,cost,'b-',](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/150355/slide-12.jpg)
![Выбор уровня [mincost,minloc] = min(cost); cutoff = mincost + secost(minloc);](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/150355/slide-13.jpg)
 Создание и обработка изображений средствами Adobe Photoshop (обзор возможностей)
Создание и обработка изображений средствами Adobe Photoshop (обзор возможностей) Хімічна інформатика
Хімічна інформатика Analysis and Design of Data Systems. Enhanced ER (EER) Mode. (Lecture 11)
Analysis and Design of Data Systems. Enhanced ER (EER) Mode. (Lecture 11) История создания UNIX-систем. (Занятия 3 и 4)
История создания UNIX-систем. (Занятия 3 и 4) Реферативная база данных
Реферативная база данных Операционные системы. Управление процессами
Операционные системы. Управление процессами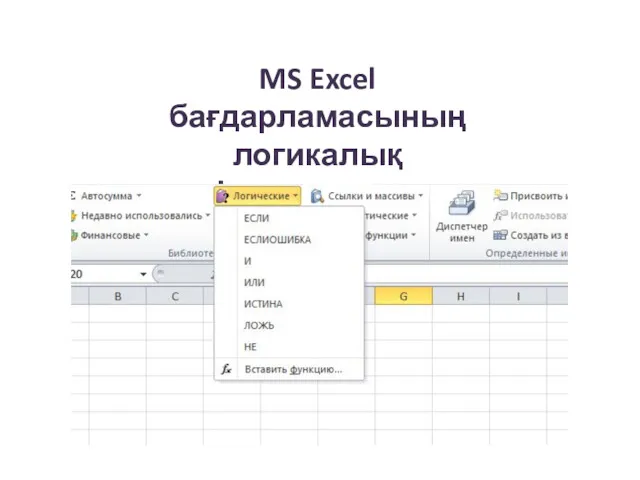 MS Excel бағдарламасының логикалық функциялары
MS Excel бағдарламасының логикалық функциялары Электронное портфолио студентов
Электронное портфолио студентов Количество информации
Количество информации Исполнитель Робот. Вспомогательные алгоритмы
Исполнитель Робот. Вспомогательные алгоритмы Встроенный язык 1С. Особенности. Основные приемы работы. Основные конструкции встроенного языка. Лекция 2.1
Встроенный язык 1С. Особенности. Основные приемы работы. Основные конструкции встроенного языка. Лекция 2.1 Принципы автоматизированного управления приточно-вытяжной вентиляцией
Принципы автоматизированного управления приточно-вытяжной вентиляцией Понятие и свойства алгоритма
Понятие и свойства алгоритма Обучение SCRATCH
Обучение SCRATCH Форматирование документа
Форматирование документа Фрагментация и персонификация контента в интернете
Фрагментация и персонификация контента в интернете Компьютеризация животноводства в РФ
Компьютеризация животноводства в РФ Алгоритмы и программирование, язык Паскаль (часть 3)
Алгоритмы и программирование, язык Паскаль (часть 3) Основные понятия культура и информация
Основные понятия культура и информация Устройства ввода-вывода
Устройства ввода-вывода Программное обеспечение компьютера
Программное обеспечение компьютера Тестирование и тестировщики. Категории программных ошибок
Тестирование и тестировщики. Категории программных ошибок Сетевая этика. Культура общения в сети
Сетевая этика. Культура общения в сети Топология сети. Блок 2
Топология сети. Блок 2 Введение в дисциплину Информатика и ИКТ
Введение в дисциплину Информатика и ИКТ Правила составления библиографического списка литературы и оформления библиографических ссылок
Правила составления библиографического списка литературы и оформления библиографических ссылок Розроблення атоматизованої системи керування робочим процесом в закладах громадського харчування
Розроблення атоматизованої системи керування робочим процесом в закладах громадського харчування Алгоритмы. Алгоритм ветвления.
Алгоритмы. Алгоритм ветвления.