Содержание
- 2. CYTOGENETIC CYTOGENETICS IS ESSENTIALLY A BRANCH OF GENETICS, BUT IS ALSO A PART OF CELL BIOLOGY/CYTOLOGY
- 4. HISTORY AND EVOLUTION OF CYTOGENETICS has been a key part of biology since 1842, when Swiss
- 5. Cytogenetic methods 1 – karyotyping 2 - Fluorescent in situ hybridization
- 6. karyotyping The routine chromosome analysis (Karyotyping) refers to analysis of metaphase chromosomes which have been banded
- 8. staining The study of karyotypes is made possible by staining. Usually, a suitable dye, such as
- 10. Chromosome abnormalities CHROMOSOMAL ABNORMALITIES THAT LEAD TO DISEASE IN HUMANS INCLUDE TURNER SYNDROME RESULTS FROM A
- 11. SOME DISORDERS ARISE FROM LOSS OF JUST A PIECE OF ONE CHROMOSOME, INCLUDING Cri du chat
- 13. FLUORESCENCE IN SITU HYBRIDIZATION (FISH) IS A LABORATORY TECHNIQUE FOR DETECTING AND LOCATING A SPECIFIC DNA
- 14. LINKAGE MAPPING USING MOLECULAR MARKERS THE NEXT STEP IN GENE ID IS TO GENETICALLY MAP ITS
- 15. Linkage Mapping of Disease Gene Location by Recombination Analysis Although it is not commonly used in
- 16. Final Steps of Mutant Gene Isolation Often mutations can be mapped only to 1 cM regions
- 18. Скачать презентацию
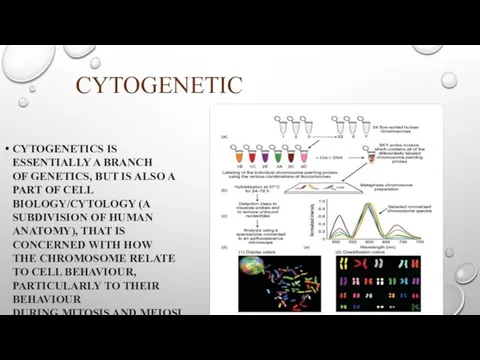
CYTOGENETIC
CYTOGENETICS IS ESSENTIALLY A BRANCH OF GENETICS, BUT IS ALSO A PART
CYTOGENETIC
CYTOGENETICS IS ESSENTIALLY A BRANCH OF GENETICS, BUT IS ALSO A PART
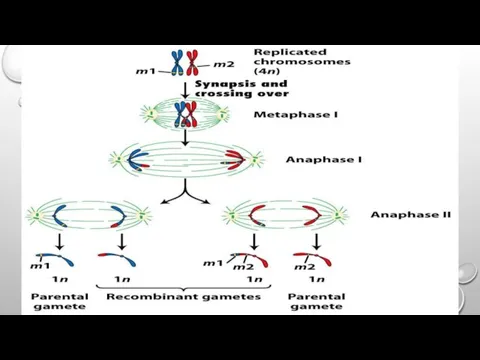
HISTORY AND EVOLUTION OF CYTOGENETICS
has been a key part of biology
HISTORY AND EVOLUTION OF CYTOGENETICS
has been a key part of biology
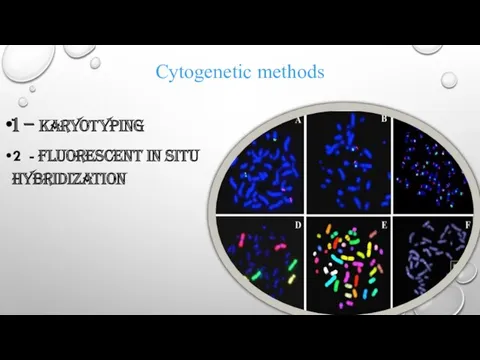
Cytogenetic methods
1 – karyotyping
2 - Fluorescent in situ hybridization
Cytogenetic methods
1 – karyotyping
2 - Fluorescent in situ hybridization
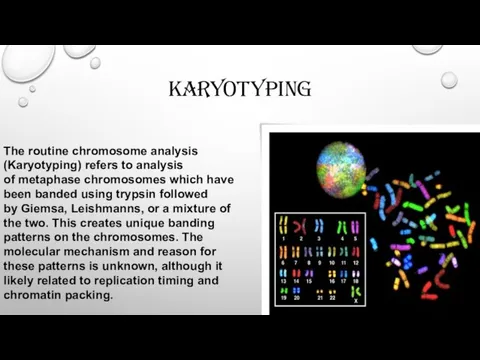
karyotyping
The routine chromosome analysis (Karyotyping) refers to analysis of metaphase chromosomes which have
karyotyping
The routine chromosome analysis (Karyotyping) refers to analysis of metaphase chromosomes which have
staining
The study of karyotypes is made possible by staining. Usually, a suitable dye,
staining
The study of karyotypes is made possible by staining. Usually, a suitable dye,
Chromosome abnormalities
CHROMOSOMAL ABNORMALITIES THAT LEAD TO DISEASE IN HUMANS INCLUDE
TURNER SYNDROME RESULTS
Chromosome abnormalities
CHROMOSOMAL ABNORMALITIES THAT LEAD TO DISEASE IN HUMANS INCLUDE
TURNER SYNDROME RESULTS
KLINEFELTER SYNDROME, THE MOST COMMON MALE CHROMOSOMAL DISEASE, OTHERWISE KNOWN AS 47,XXY, IS CAUSED BY AN EXTRA X CHROMOSOME.
EDWARDS SYNDROME IS CAUSED BY TRISOMY (THREE COPIES) OF CHROMOSOME 18.
DOWN SYNDROME, A COMMON CHROMOSOMAL DISEASE, IS CAUSED BY TRISOMY OF CHROMOSOME 21.
PATAU SYNDROME IS CAUSED BY TRISOMY OF CHROMOSOME 13.
TRISOMY 9, BELIEVED TO BE THE 4TH MOST COMMON TRISOMY, HAS MANY LONG LIVED AFFECTED INDIVIDUALS BUT ONLY IN A FORM OTHER THAN A FULL TRISOMY, SUCH AS TRISOMY 9P SYNDROME OR MOSAIC TRISOMY 9. THEY OFTEN FUNCTION QUITE WELL, BUT TEND TO HAVE TROUBLE WITH SPEECH.
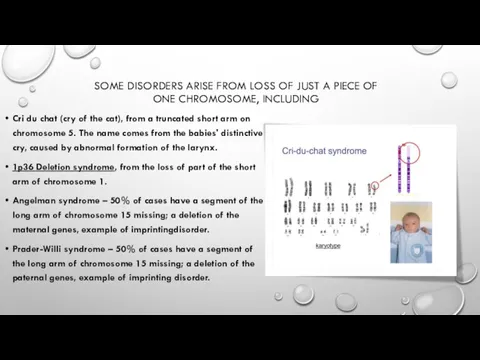
SOME DISORDERS ARISE FROM LOSS OF JUST A PIECE OF ONE
SOME DISORDERS ARISE FROM LOSS OF JUST A PIECE OF ONE
Cri du chat (cry of the cat), from a truncated short arm on chromosome 5. The name comes from the babies' distinctive cry, caused by abnormal formation of the larynx.
1p36 Deletion syndrome, from the loss of part of the short arm of chromosome 1.
Angelman syndrome – 50% of cases have a segment of the long arm of chromosome 15 missing; a deletion of the maternal genes, example of imprintingdisorder.
Prader-Willi syndrome – 50% of cases have a segment of the long arm of chromosome 15 missing; a deletion of the paternal genes, example of imprinting disorder.
FLUORESCENCE IN SITU HYBRIDIZATION (FISH) IS A LABORATORY TECHNIQUE FOR DETECTING
FLUORESCENCE IN SITU HYBRIDIZATION (FISH) IS A LABORATORY TECHNIQUE FOR DETECTING
LINKAGE MAPPING USING MOLECULAR MARKERS
THE NEXT STEP IN GENE ID IS
LINKAGE MAPPING USING MOLECULAR MARKERS
THE NEXT STEP IN GENE ID IS
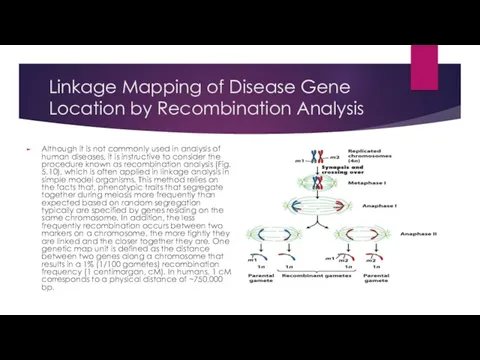
Linkage Mapping of Disease Gene Location by Recombination Analysis
Although it is
Linkage Mapping of Disease Gene Location by Recombination Analysis
Although it is
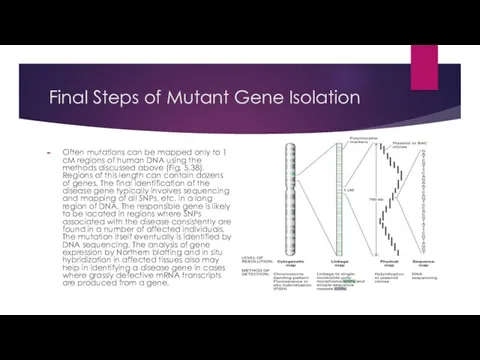
Final Steps of Mutant Gene Isolation
Often mutations can be mapped only
Final Steps of Mutant Gene Isolation
Often mutations can be mapped only








 Улучшение качества материнской и неонатальной помощи. (Модуль 15)
Улучшение качества материнской и неонатальной помощи. (Модуль 15) Синдром легочной диссеминации. Диссеминированный туберкулез легких
Синдром легочной диссеминации. Диссеминированный туберкулез легких Нейроофтальмологические нарушения при нейромышечных заболеваниях
Нейроофтальмологические нарушения при нейромышечных заболеваниях Острые пневмонии у детей
Острые пневмонии у детей Приклади хвороби органів ШКТ
Приклади хвороби органів ШКТ Введение в фитотерапию
Введение в фитотерапию Ранняя диагностика острого лейкоза у детей
Ранняя диагностика острого лейкоза у детей Синдром диабетической стопы
Синдром диабетической стопы О вреде абортов
О вреде абортов Рак слизистой оболочки полости рта. Клиника, диагностика, лечение
Рак слизистой оболочки полости рта. Клиника, диагностика, лечение Семиотика заболеваний мочевой системы у детей
Семиотика заболеваний мочевой системы у детей Автоматические поточные линии
Автоматические поточные линии ЭКГ
ЭКГ Педиатрия как наука о здоровом и больном ребёнке.Место педиатрии в системе общей медицины. Возрастная периодизация в педиатрии
Педиатрия как наука о здоровом и больном ребёнке.Место педиатрии в системе общей медицины. Возрастная периодизация в педиатрии Острая кишечная непроходимость у детей
Острая кишечная непроходимость у детей Гестационная трофобластическая болезнь
Гестационная трофобластическая болезнь Медицина в Средневековье
Медицина в Средневековье Кровообращение и гомеостаз
Кровообращение и гомеостаз Эпидемиология РМЖ
Эпидемиология РМЖ Метаболический синдром и НАЖБП в современных условиях. Актуальность коморбидности
Метаболический синдром и НАЖБП в современных условиях. Актуальность коморбидности Гигиена аптечных заведений
Гигиена аптечных заведений Клубные наркотики
Клубные наркотики Логопедия. Дизартрия
Логопедия. Дизартрия Координаторная сфера. Мозжечок. Синдромы поражения
Координаторная сфера. Мозжечок. Синдромы поражения Орально-мануальные техники
Орально-мануальные техники Анатомо-физиологические особенности лёгких. (Лекция 11)
Анатомо-физиологические особенности лёгких. (Лекция 11) Общественное здоровье-высшая ценность человечества
Общественное здоровье-высшая ценность человечества Ерте жастағы балалардағы сөйлеу тілінің тежелуінің алдын алудың ерекшеліктері
Ерте жастағы балалардағы сөйлеу тілінің тежелуінің алдын алудың ерекшеліктері