Содержание
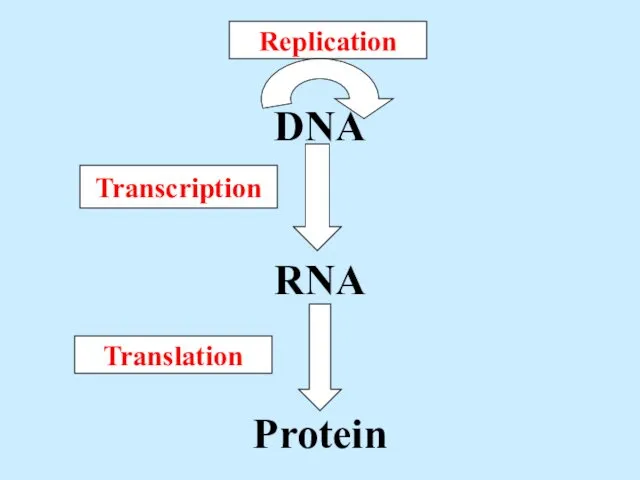
- 2. DNA RNA Protein Replication Transcription Translation
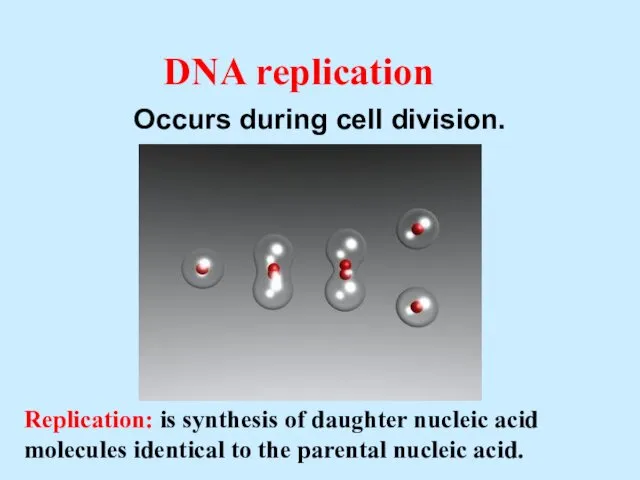
- 3. DNA replication Occurs during cell division. Replication: is synthesis of daughter nucleic acid molecules identical to
- 4. Replication of the DNA proceeds in stages: Initiation Elongation Termination DNA replication requires many enzymes and
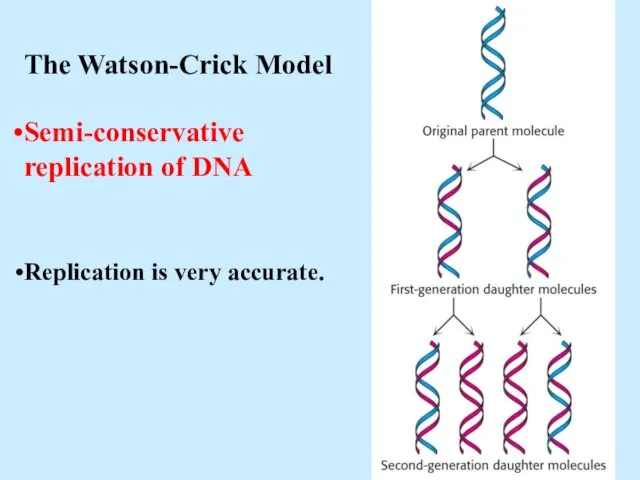
- 5. The Watson-Crick Model Semi-conservative replication of DNA Replication is very accurate.
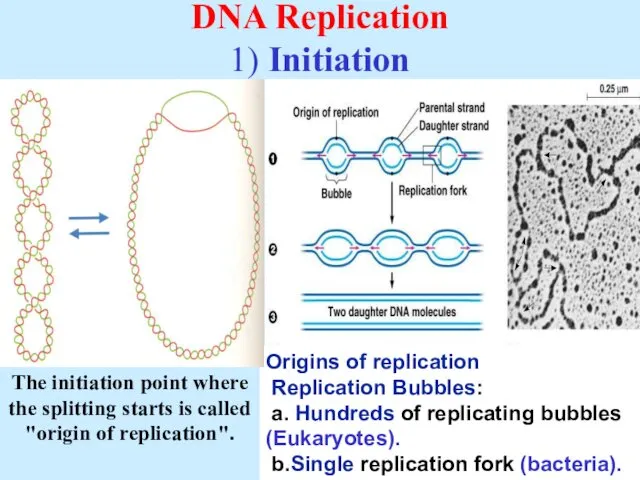
- 6. DNA Replication 1) Initiation Origins of replication Replication Bubbles: a. Hundreds of replicating bubbles (Eukaryotes). b.Single
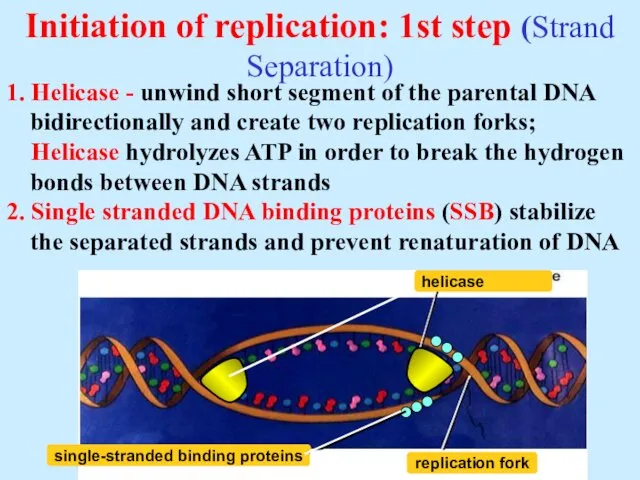
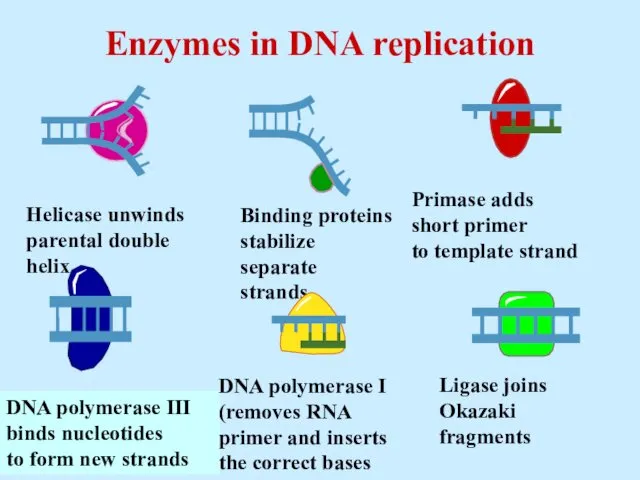
- 7. Initiation of replication: 1st step (Strand Separation) 1. Helicase - unwind short segment of the parental
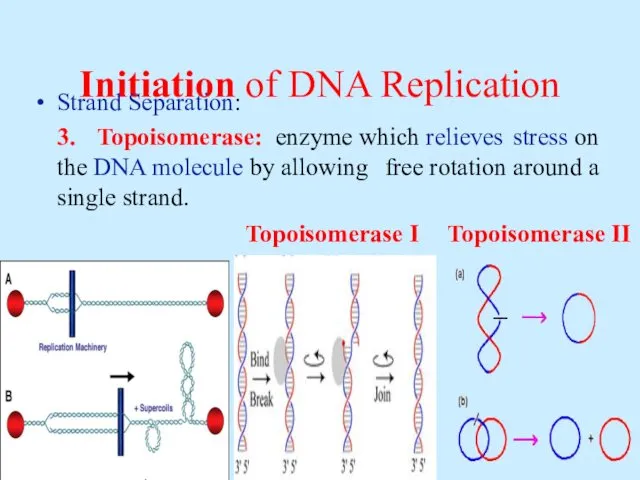
- 8. Initiation of DNA Replication Strand Separation: 3. Topoisomerase: enzyme which relieves stress on the DNA molecule
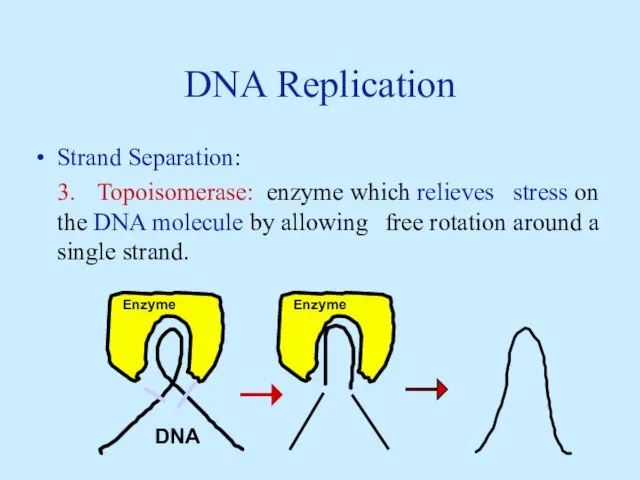
- 9. DNA Replication Strand Separation: 3. Topoisomerase: enzyme which relieves stress on the DNA molecule by allowing
- 10. 2. Elongation - Both Template strands are copied at a Replication Fork DNA replication is cataly
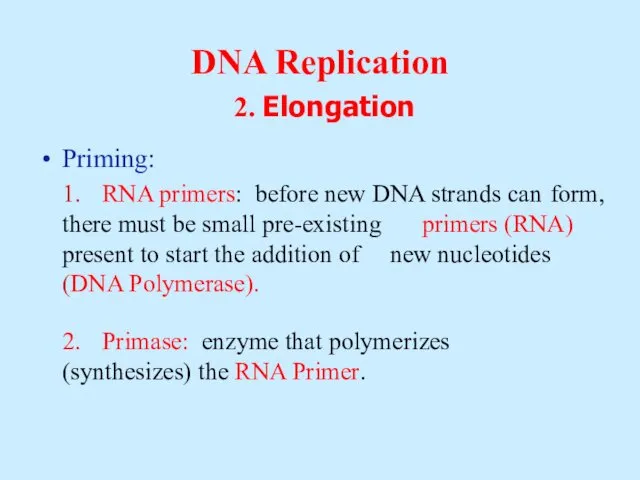
- 11. DNA Replication 2. Elongation Priming: 1. RNA primers: before new DNA strands can form, there must
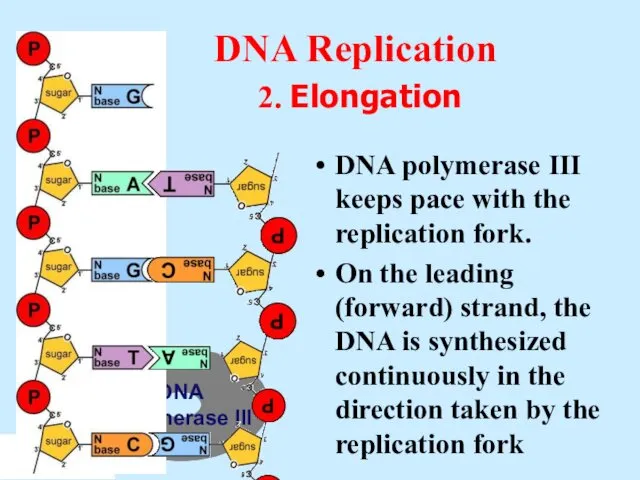
- 12. DNA Polymerase III DNA Replication 2. Elongation DNA polymerase III keeps pace with the replication fork.
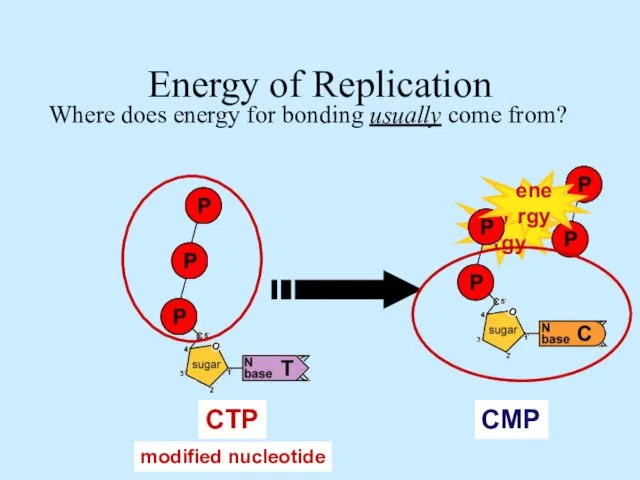
- 13. energy ATP GTP TTP CTP Energy of Replication Where does energy for bonding usually come from?
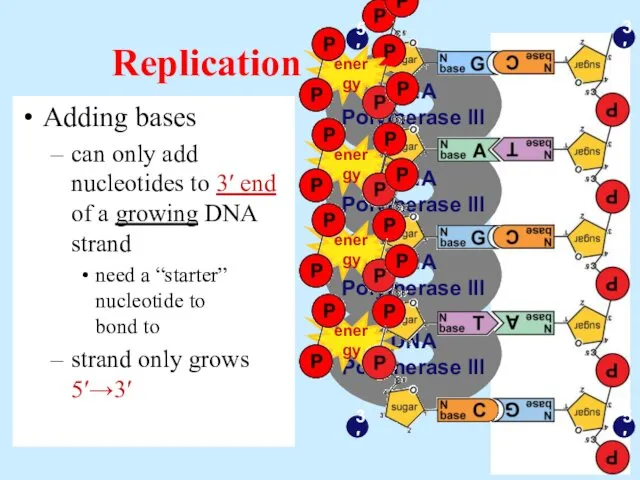
- 14. Adding bases can only add nucleotides to 3′ end of a growing DNA strand need a
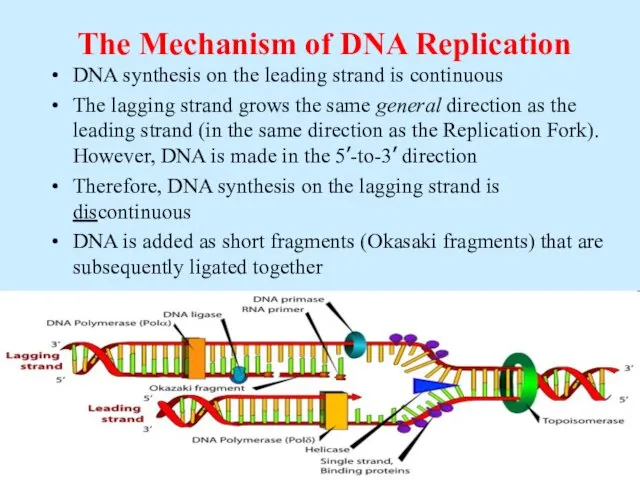
- 15. The Mechanism of DNA Replication DNA synthesis on the leading strand is continuous The lagging strand
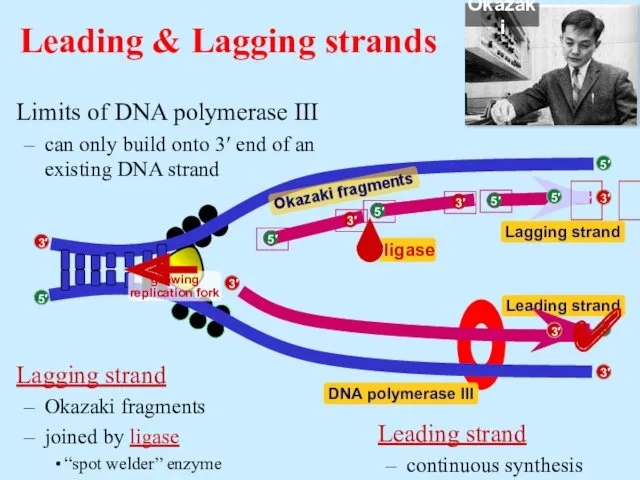
- 16. Limits of DNA polymerase III can only build onto 3′ end of an existing DNA strand
- 17. Replication fork / Replication bubble leading strand lagging strand leading strand lagging strand leading strand lagging
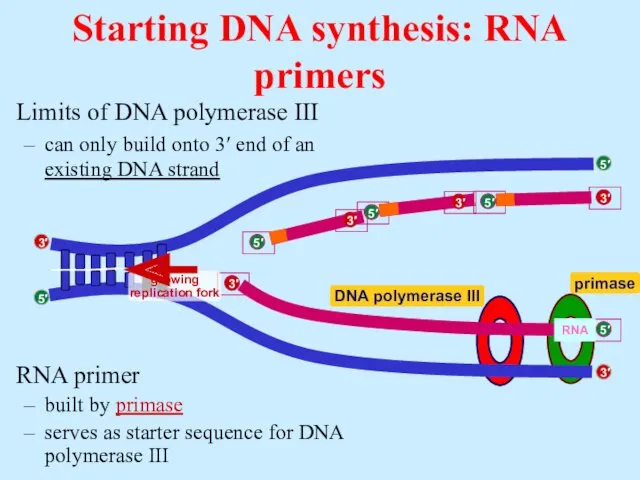
- 18. RNA primer built by primase serves as starter sequence for DNA polymerase III Limits of DNA
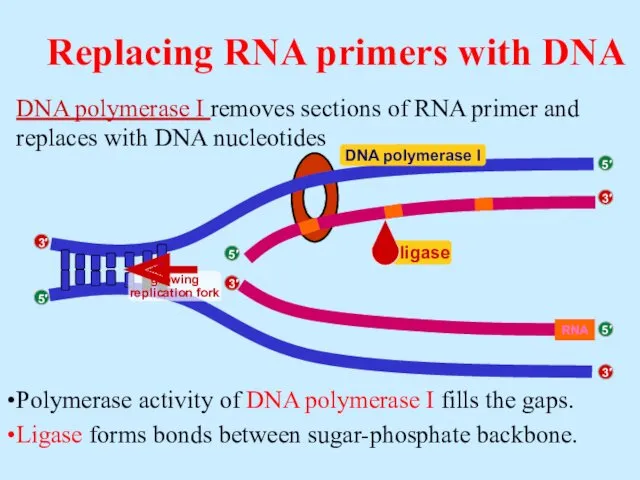
- 19. DNA polymerase I removes sections of RNA primer and replaces with DNA nucleotides Polymerase activity of
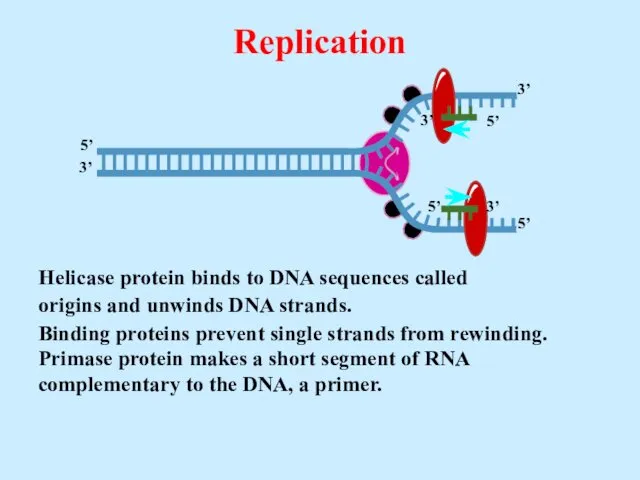
- 20. Helicase protein binds to DNA sequences called origins and unwinds DNA strands. Replication
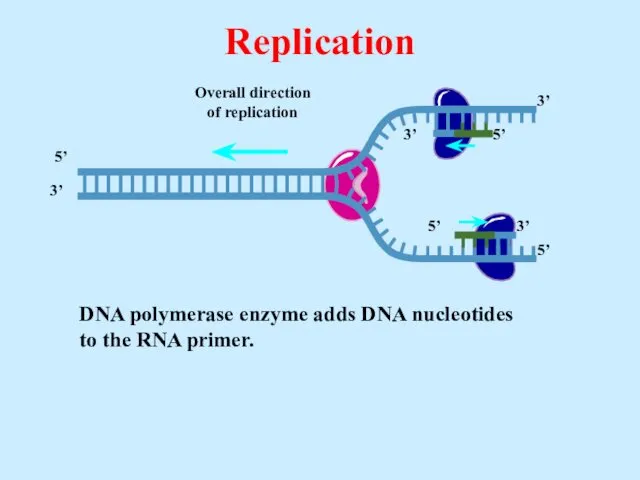
- 21. DNA polymerase enzyme adds DNA nucleotides to the RNA primer. Replication
- 22. DNA polymerase enzyme adds DNA nucleotides to the RNA primer. DNA polymerase proofreads bases added and
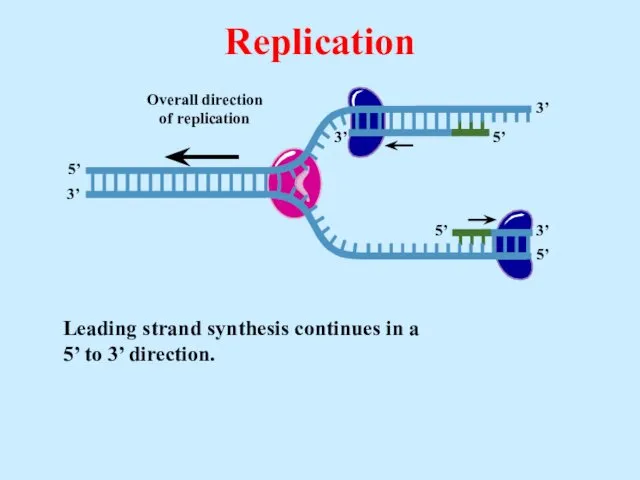
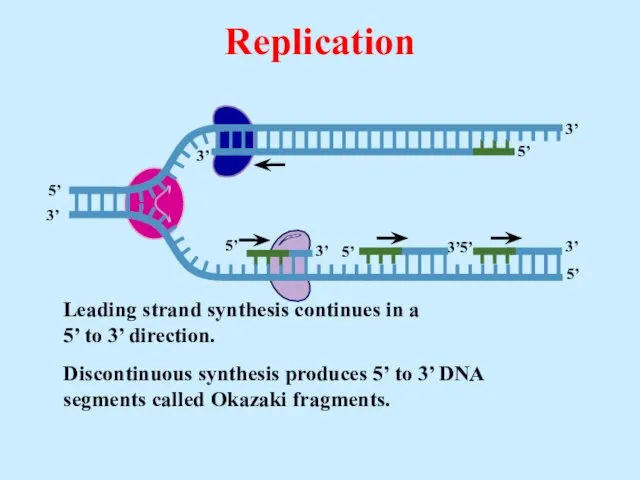
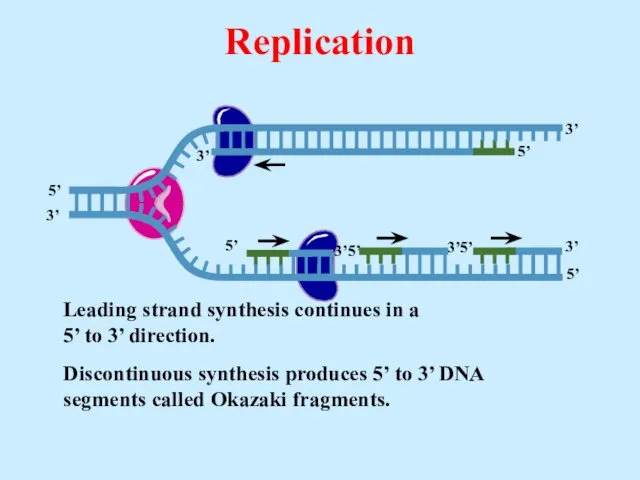
- 23. Leading strand synthesis continues in a 5’ to 3’ direction. Replication
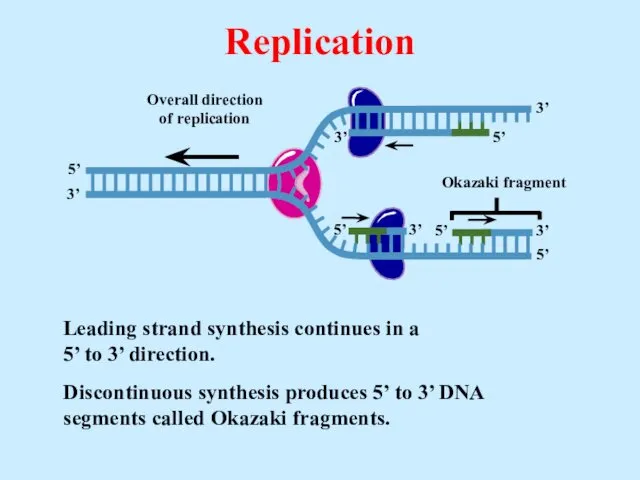
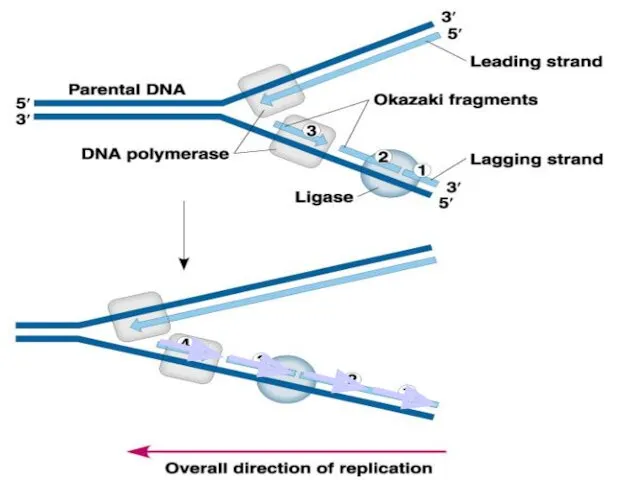
- 24. Leading strand synthesis continues in a 5’ to 3’ direction. Discontinuous synthesis produces 5’ to 3’
- 25. 5’ 5’ 5’ 3’ 5’ 3’ 3’ 5’ 3’ Overall direction of replication 3’ Leading strand
- 26. 5’ 5’ 3’ 5’ 3’ 3’ 5’ 3’ 3’ 5’ 5’ 3’ Leading strand synthesis continues
- 27. 3’ 5’ 3’ Leading strand synthesis continues in a 5’ to 3’ direction. Discontinuous synthesis produces
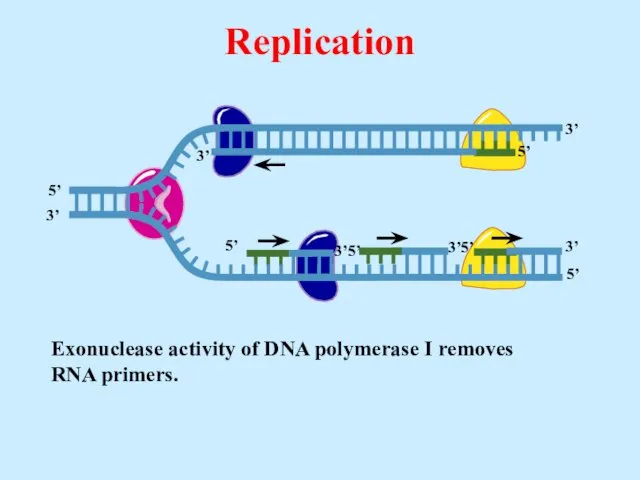
- 28. Exonuclease activity of DNA polymerase I removes RNA primers. Replication
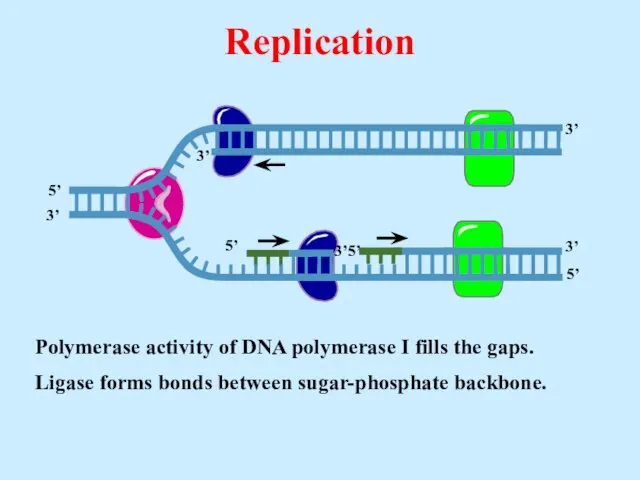
- 29. Polymerase activity of DNA polymerase I fills the gaps. Ligase forms bonds between sugar-phosphate backbone. Replication
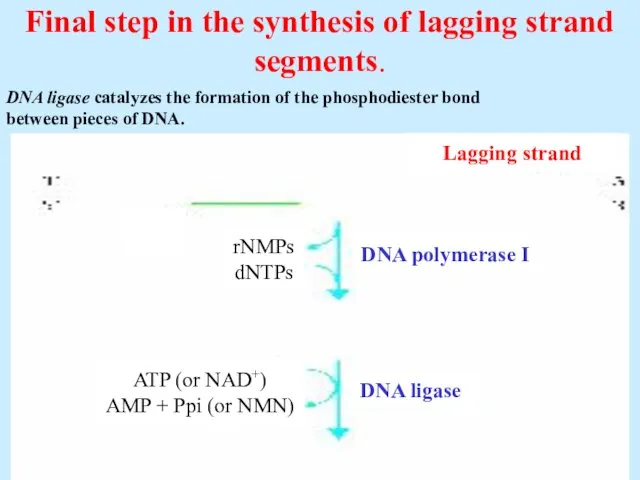
- 31. Final step in the synthesis of lagging strand segments. Lagging strand rNMPs dNTPs ATP (or NAD+)
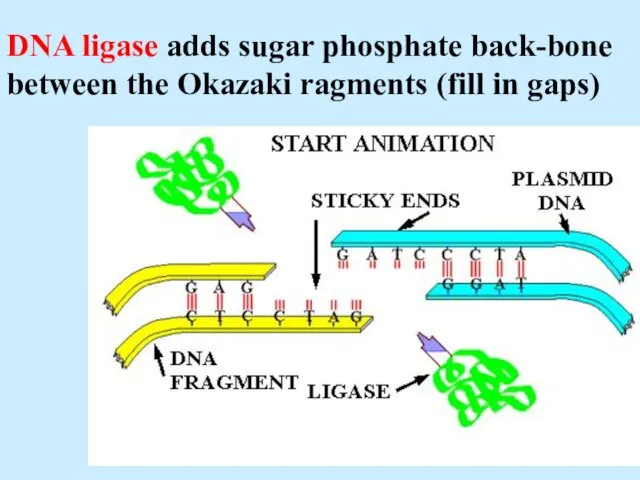
- 32. DNA ligase adds sugar phosphate back-bone between the Okazaki ragments (fill in gaps)
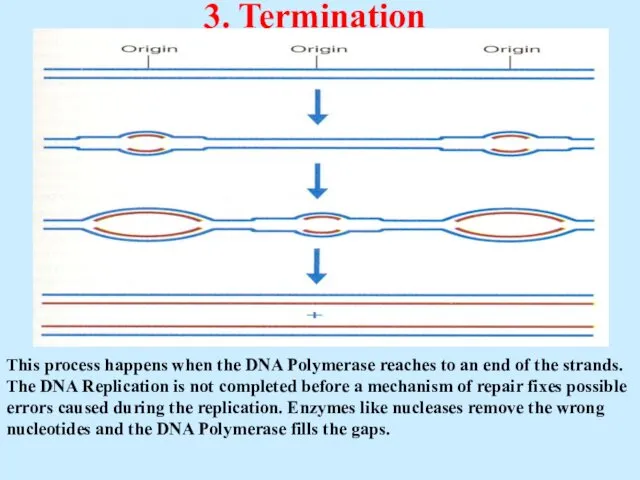
- 33. 3. Termination This process happens when the DNA Polymerase reaches to an end of the strands.
- 34. Enzymes in DNA replication
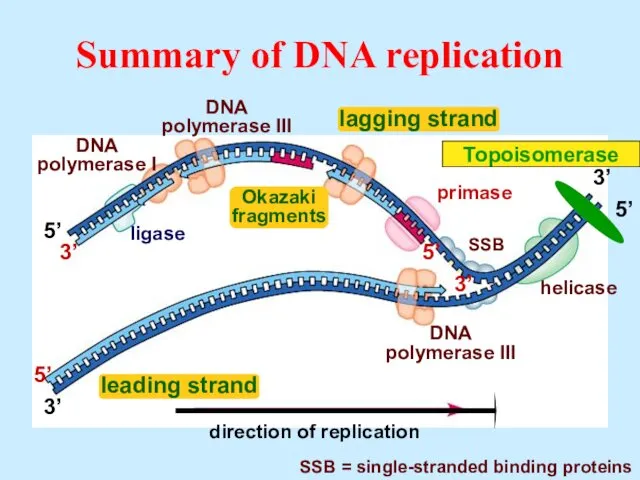
- 35. Summary of DNA replication 3’ 5’ 3’ 5’ 5’ 3’ 3’ 5’ helicase SSB = single-stranded
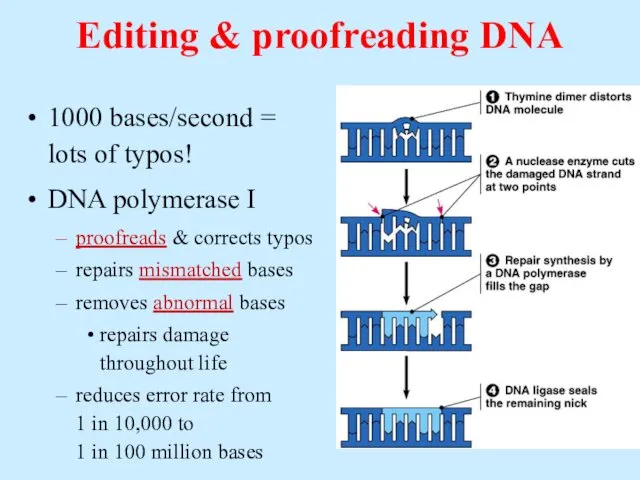
- 36. Editing & proofreading DNA 1000 bases/second = lots of typos! DNA polymerase I proofreads & corrects
- 38. Part I Protein sinthesis
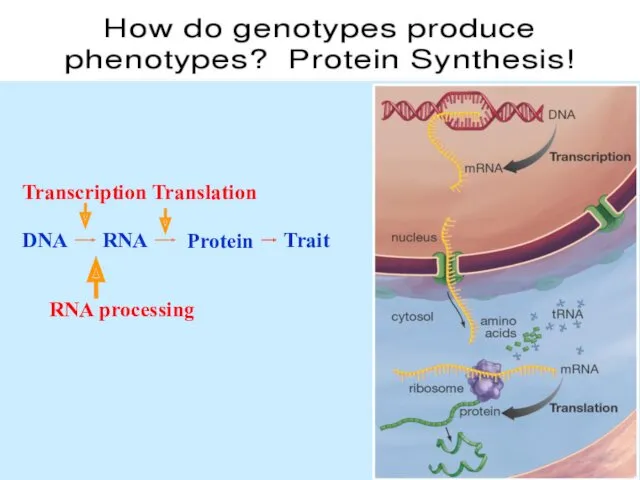
- 39. DNA Trait RNA Protein The “Central Dogma” of Molecular Genetics
- 40. The chemical nature of RNA differs from that of DNA. Ribonucleic acid (RNA) is a polymer
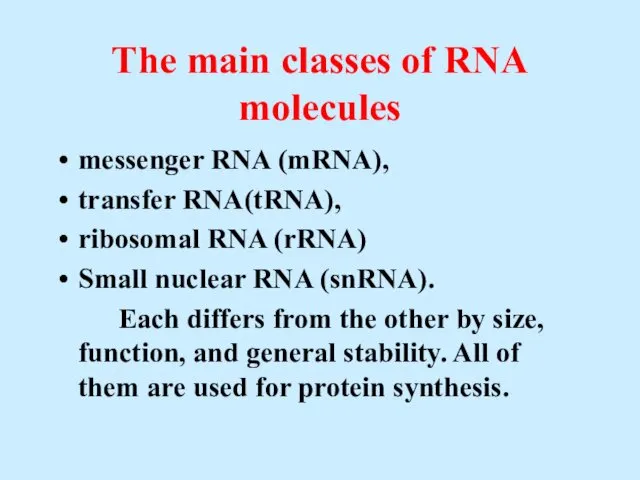
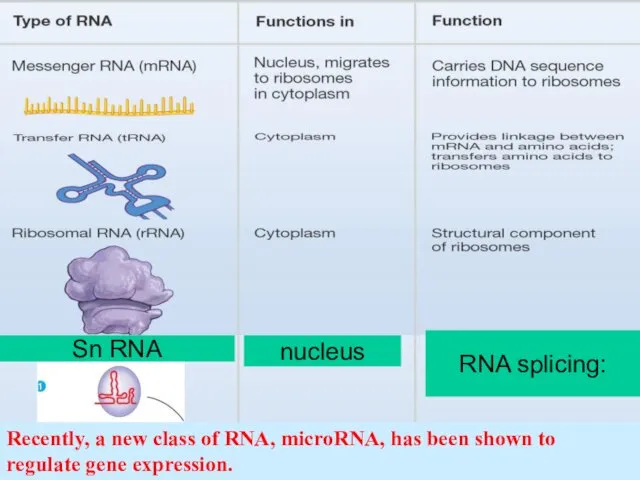
- 41. The main classes of RNA molecules messenger RNA (mRNA), transfer RNA(tRNA), ribosomal RNA (rRNA) Small nuclear
- 42. Recently, a new class of RNA, microRNA, has been shown to regulate gene expression. nucleus Sn
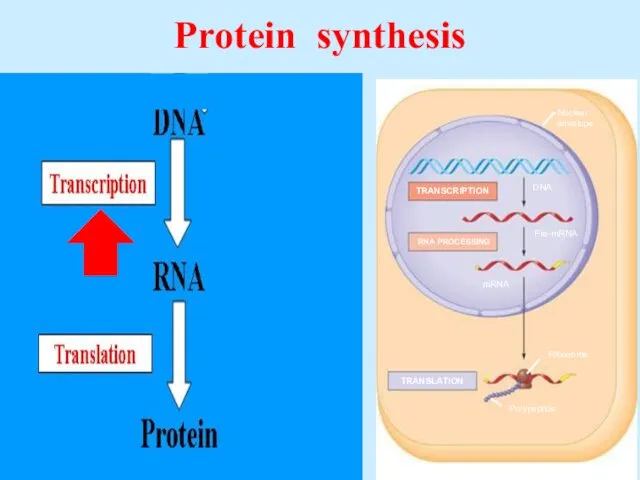
- 43. Protein synthesis
- 44. Transcription is DNA-dependent synthesis of RNA. Transcription is catalyzed by RNA polymerase. RNA polymerase copies a
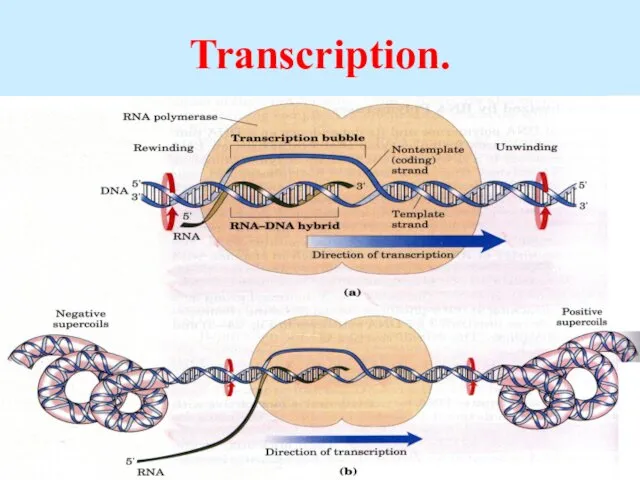
- 45. Transcription.
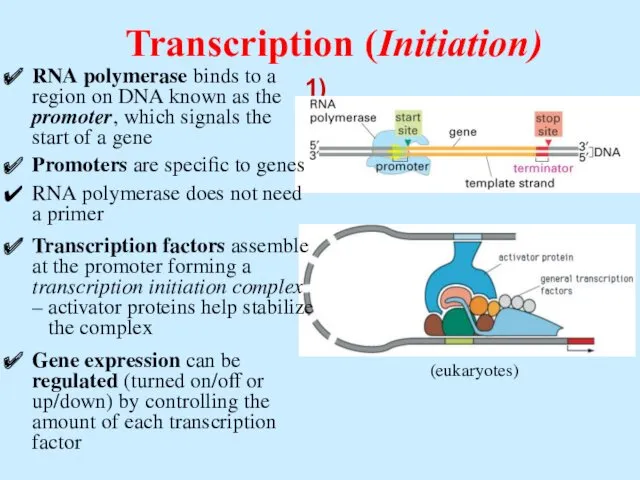
- 46. Transcription (Initiation) RNA polymerase binds to a region on DNA known as the promoter, which signals
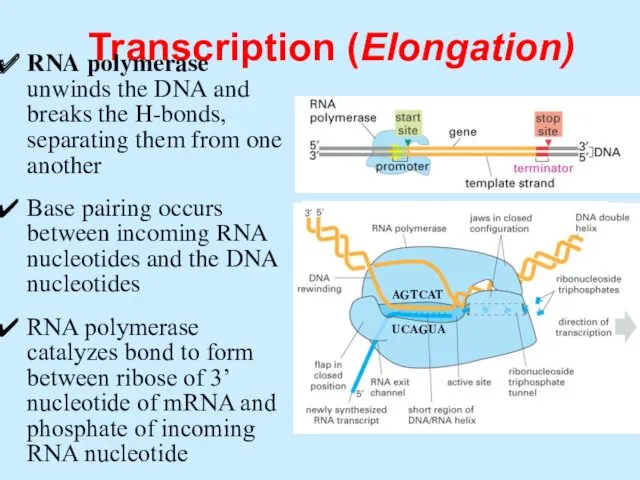
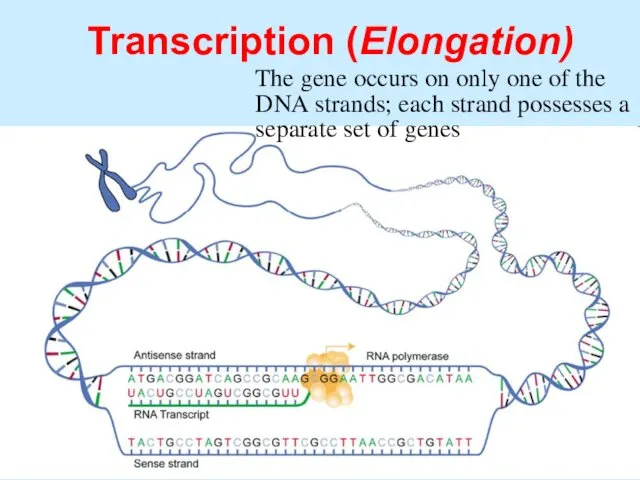
- 47. Transcription (Elongation) RNA polymerase unwinds the DNA and breaks the H-bonds, separating them from one another
- 48. Transcription (Elongation) The gene occurs on only one of the DNA strands; each strand possesses a
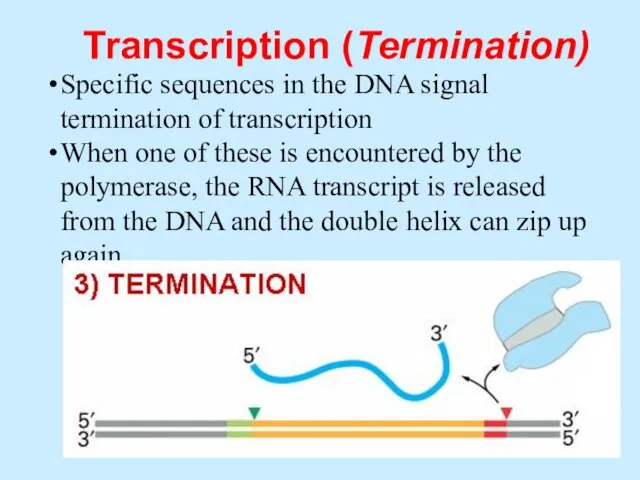
- 49. Transcription (Termination) Specific sequences in the DNA signal termination of transcription When one of these is
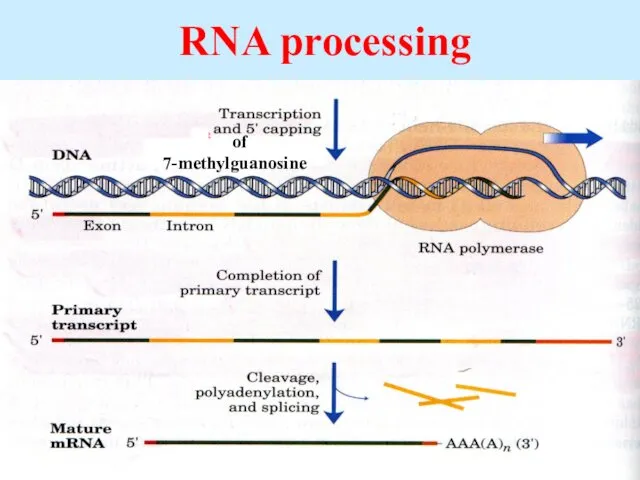
- 50. RNA processing of 7-methylguanosine
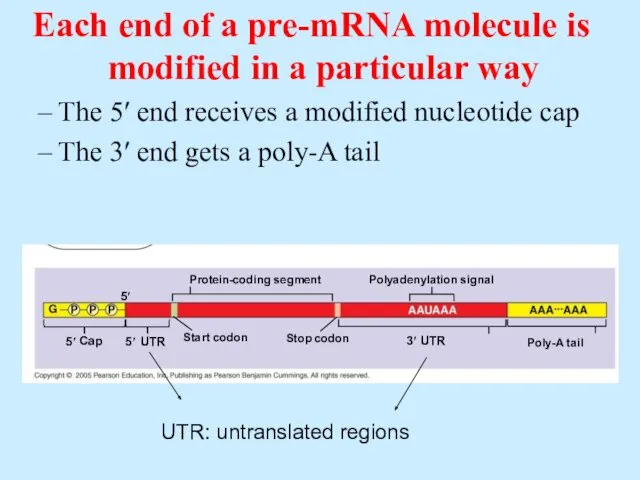
- 51. 5′ Protein-coding segment 5′ Start codon Stop codon Poly-A tail Polyadenylation signal 5′ 3′ Cap UTR
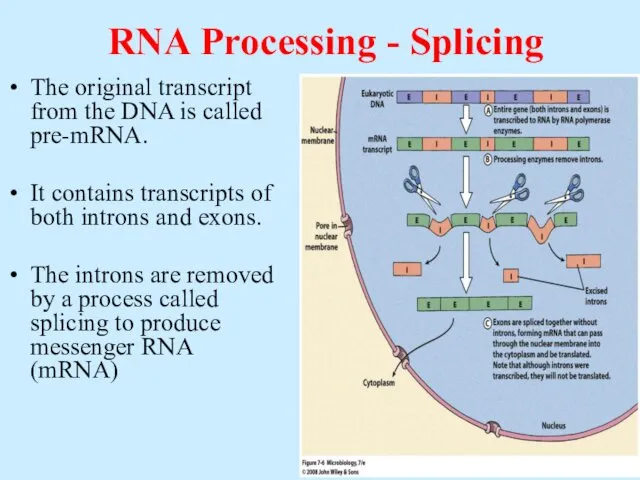
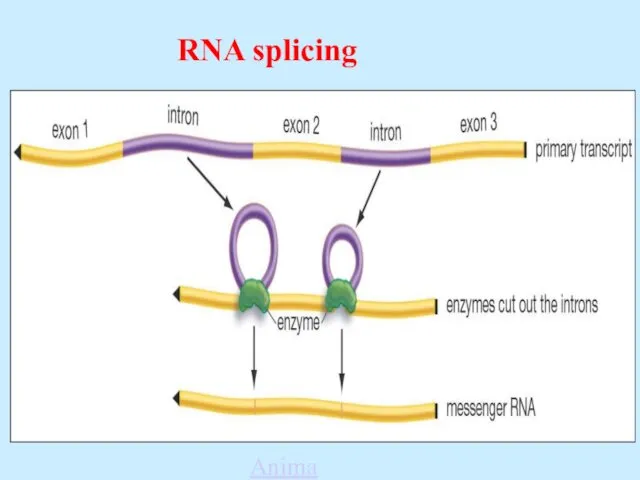
- 52. RNA Processing - Splicing The original transcript from the DNA is called pre-mRNA. It contains transcripts
- 53. Spliceosomes - complex of proteins and several small nuclear ribonucleoproteins (snRNPs) Recognize splice sites (specific RNA
- 54. RNA splicing Animation
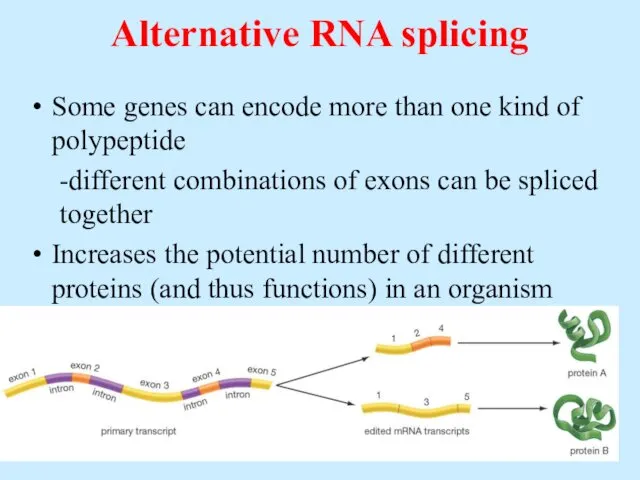
- 55. Alternative RNA splicing Some genes can encode more than one kind of polypeptide -different combinations of
- 57. Скачать презентацию







 Сходства и различия клеток растений, животных и грибов
Сходства и различия клеток растений, животных и грибов Использование дрожжевых грибов для производства разных групп продуктов
Использование дрожжевых грибов для производства разных групп продуктов Открытое мероприятие по профилактике вредных привычек
Открытое мероприятие по профилактике вредных привычек Бактерії - прокаріотичні одноклітинні мікроорганізми
Бактерії - прокаріотичні одноклітинні мікроорганізми Влияние ГМО на организм человека
Влияние ГМО на организм человека Общая физиология сенсорных систем
Общая физиология сенсорных систем Половое поведение животных
Половое поведение животных Дыхание растений
Дыхание растений Круговорот кислорода в природе
Круговорот кислорода в природе Звёздный час. Загадки о животных и растениях
Звёздный час. Загадки о животных и растениях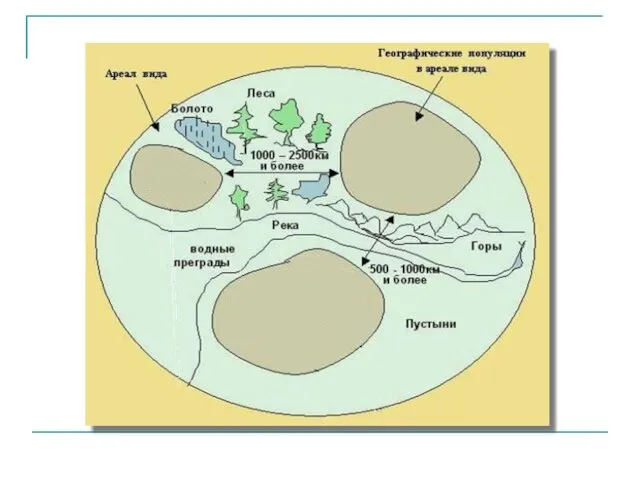 Популяция. Популяционная генетика
Популяция. Популяционная генетика Сахароза. Дисахариды (олигосахариды)
Сахароза. Дисахариды (олигосахариды) Клетки и их разнообразие в многоклеточном организме
Клетки и их разнообразие в многоклеточном организме Основы гистологии. Виды тканей
Основы гистологии. Виды тканей An introduction to metabolism
An introduction to metabolism Глюкоза. Молекулярна формула. Фізичні та хімічні властивості глюкози. Поширення в природі
Глюкоза. Молекулярна формула. Фізичні та хімічні властивості глюкози. Поширення в природі 20231101_vestniki_vesny_viktorina
20231101_vestniki_vesny_viktorina Глаз
Глаз Cattle breeds
Cattle breeds Как зимой помочь птицам (урок окружающего мира, 1 класс)
Как зимой помочь птицам (урок окружающего мира, 1 класс) Эффективный способ посадки картофеля
Эффективный способ посадки картофеля Мы говорим Нет алкоголю!
Мы говорим Нет алкоголю! Физиология растений. Механизмы поступления воды в растительную клетку. Дальний транспорт воды в растении
Физиология растений. Механизмы поступления воды в растительную клетку. Дальний транспорт воды в растении Презентация по биологии для 9 класса по теме:Метаболизм. Энергетический обмен.
Презентация по биологии для 9 класса по теме:Метаболизм. Энергетический обмен. Животные Антарктиды
Животные Антарктиды Красная книга Удмуртии. Животные
Красная книга Удмуртии. Животные Кератин в волосах и восстановление волос
Кератин в волосах и восстановление волос Катархей. Архей
Катархей. Архей