Содержание
- 2. PHYLOGENETIC DISORDERS OF RESPIRATORY SYSTEM REPRESENTED BY : DHRUV MANGAL 195 b (LA-2) SUPERVISOR: ANNA ZHUKOVA
- 3. Respiratory syncytial virus (RSV) is a major cause of lower respiratory tract infection in young children
- 4. Respiratory syncytial virus (RSV) is an important cause of bronchiolitis and pneumonia in infants and young
- 5. A commercial multiplex PCR assay (Seeplex RV7, Seegene, Seoul, South Korea) was used to screen for
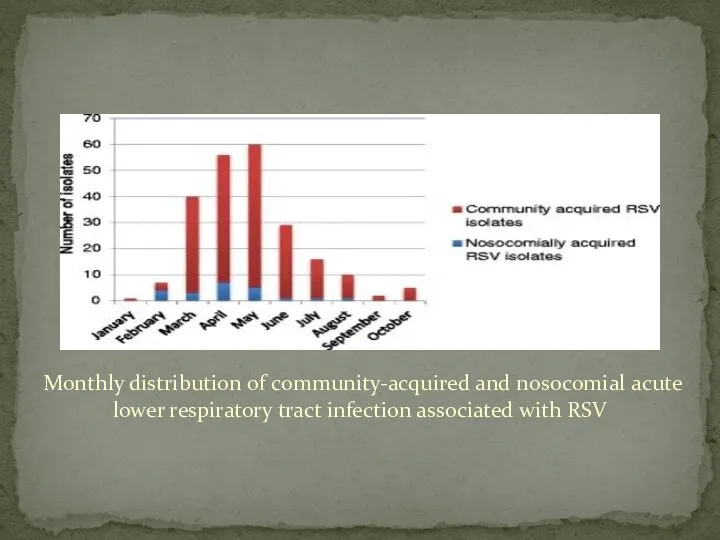
- 6. Two-hundred and twenty-six children with PCR-confirmed RSV acute lower respiratory tract infection were identified during the
- 7. The median duration of symptoms preceding hospitalisation was 2 days (IQR: 1–4 days). As shown in
- 8. RSV A and RSV B accounted for 181 (80.1 %) and 45 (19.9 %) of the
- 9. Monthly distribution of community-acquired and nosocomial acute lower respiratory tract infection associated with RSV
- 10. Factors significantly associated with nosocomial infection on univariate analysis included age 6 months or older and
- 11. In order to evaluate the circulation of the different human rhinovirus (HRV) species and genotypes in
- 12. The study was carried out in Pediatric Clinic 1 of the Department of Pathophysiology and Transplantation
- 13. Viral nucleic acids were extracted from the nasopharyngeal swabs using a Nuclisens EasyMAG automated extraction system
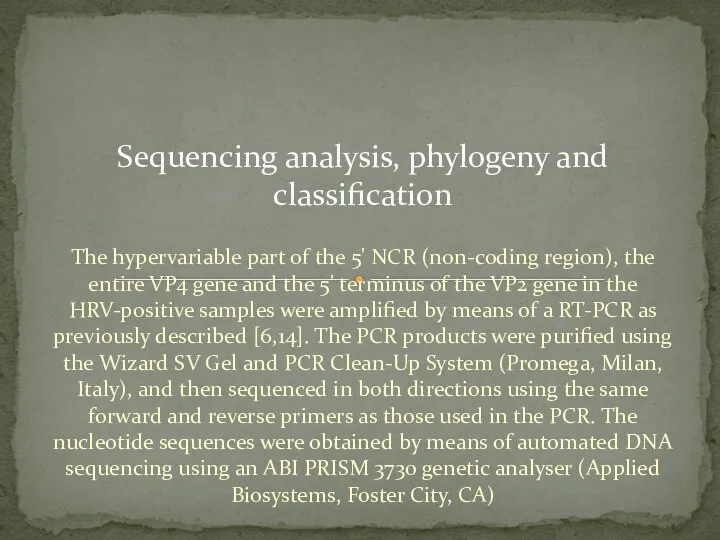
- 14. The hypervariable part of the 5' NCR (non-coding region), the entire VP4 gene and the 5'
- 15. The HRV sequences showed marked genetic diversity. The HRV-C sequences were the most heterogenous, with an
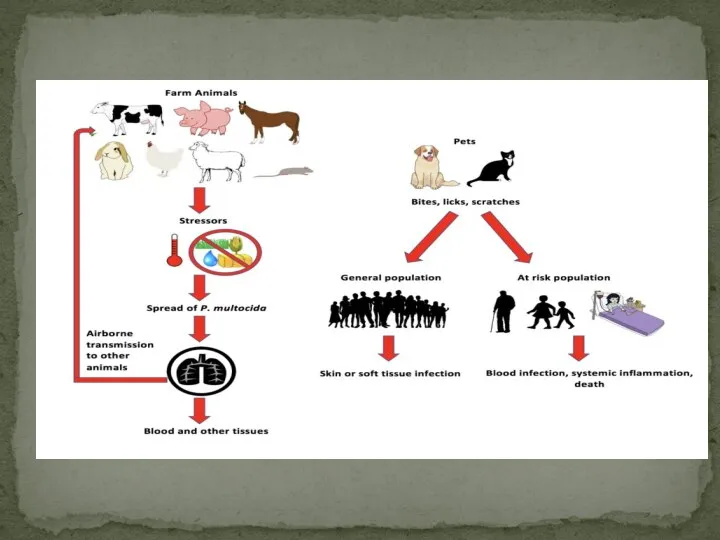
- 16. Pasteurella multocida is a leading cause of respiratory diseases in many host species. To understand the
- 17. A total of 47 P. multocida strains were selected for whole genome sequencing in this study
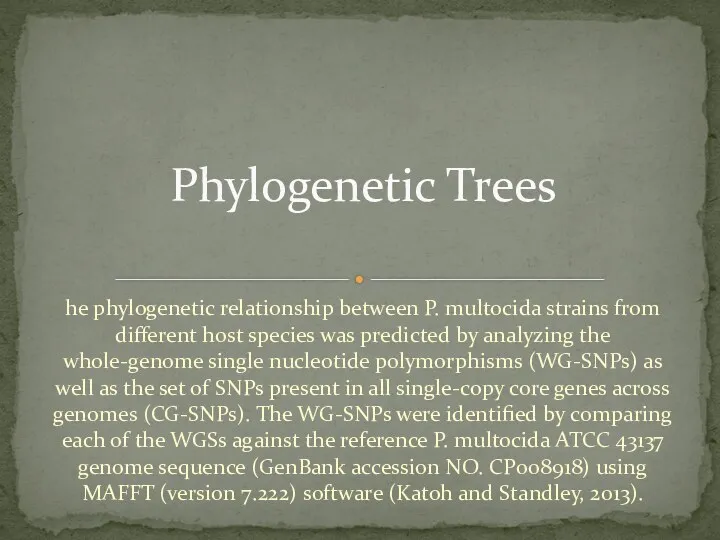
- 18. he phylogenetic relationship between P. multocida strains from different host species was predicted by analyzing the
- 20. Whole genome sequencing yielded approximately 796.25~1823.87 Mbp raw data for the 47 porcine P. multocida isolates.
- 22. Скачать презентацию












 Общие правила оказания первой доврачебной помощи. Алгоритм оказания первой помощи. Юридические и моральные аспекты
Общие правила оказания первой доврачебной помощи. Алгоритм оказания первой помощи. Юридические и моральные аспекты Клинические формы вторичного туберкулеза
Клинические формы вторичного туберкулеза Рентгеноконтрастные исследования и препараты
Рентгеноконтрастные исследования и препараты Анатомо-физиологические особенности эндокринной системы у детей
Анатомо-физиологические особенности эндокринной системы у детей Мочекаменная болезнь. Гидронефроз
Мочекаменная болезнь. Гидронефроз Вич и Спид
Вич и Спид Інфузійна терапія
Інфузійна терапія Градация доказательств и уровни рекомендаций
Градация доказательств и уровни рекомендаций Особенности сестринского ухода за инфекционными больными. Сестринский процесс. Сестринский диагноз
Особенности сестринского ухода за инфекционными больными. Сестринский процесс. Сестринский диагноз Анализ затрат на лекарственные средства с помощью ABC/VEV методологии
Анализ затрат на лекарственные средства с помощью ABC/VEV методологии Моногибридті будандастыру. Гибридологиялық зерттеу әдісі
Моногибридті будандастыру. Гибридологиялық зерттеу әдісі Опухоли. Онкология
Опухоли. Онкология Анонимные Наркоманы г. Йошкар-Ола
Анонимные Наркоманы г. Йошкар-Ола Энтеробиоз: определение
Энтеробиоз: определение Ожирение. Степени ожирения
Ожирение. Степени ожирения Генетика человека. Генные болезни
Генетика человека. Генные болезни Острые воспалительные заболевания матки и придатков как причина развития клиники острого живота в гинекологии
Острые воспалительные заболевания матки и придатков как причина развития клиники острого живота в гинекологии Лекарственная токсикология
Лекарственная токсикология Орталық және шеткі жүйке жүйесінің клиникалық физиологиясы бен биохимиясы
Орталық және шеткі жүйке жүйесінің клиникалық физиологиясы бен биохимиясы История развития психогенетики в мировой науке
История развития психогенетики в мировой науке Ас қорыту жүйесіне жалпы шолу
Ас қорыту жүйесіне жалпы шолу Микробиологическая диагностика брюшного тифа, паратифов и других сальмонеллезных инфекций. Пищевые отравления и их диагностика
Микробиологическая диагностика брюшного тифа, паратифов и других сальмонеллезных инфекций. Пищевые отравления и их диагностика Лабораторная диагностика заболеваний, вызываемых извитыми формами бактерий. Спирохетозы (сифилис, лептоспироз, возвратные тифы)
Лабораторная диагностика заболеваний, вызываемых извитыми формами бактерий. Спирохетозы (сифилис, лептоспироз, возвратные тифы) Жарақаттар
Жарақаттар Диагностическая информативность онкомаркеров в гинекологии
Диагностическая информативность онкомаркеров в гинекологии Специфическая (антидотная) фармакотерапия острых отравлений
Специфическая (антидотная) фармакотерапия острых отравлений Эректильная дисфункция (ЭД)
Эректильная дисфункция (ЭД) Лекарственные препараты по химии
Лекарственные препараты по химии