Содержание
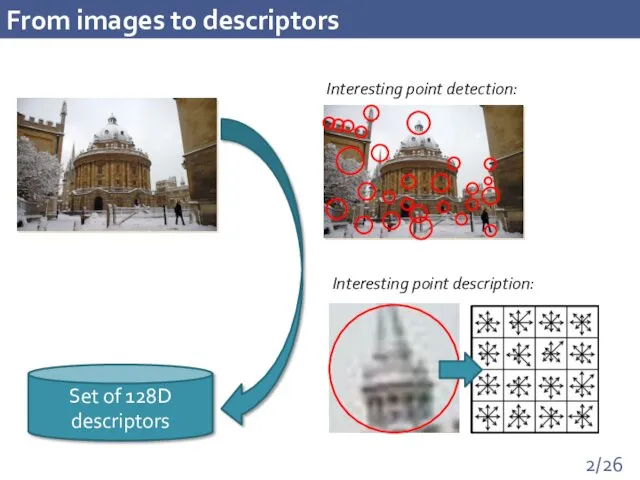
- 2. From images to descriptors
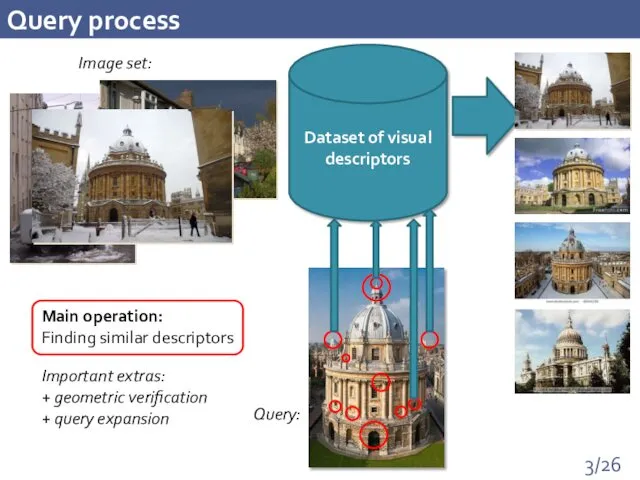
- 3. Query process Dataset of visual descriptors Image set: Query: Important extras: + geometric verification + query
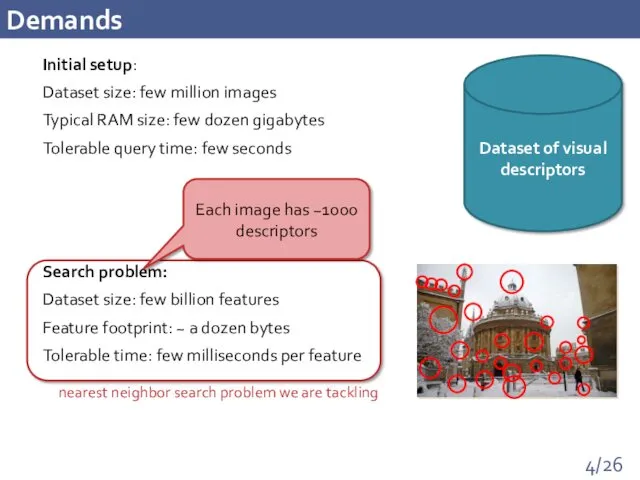
- 4. Demands Initial setup: Dataset size: few million images Typical RAM size: few dozen gigabytes Tolerable query
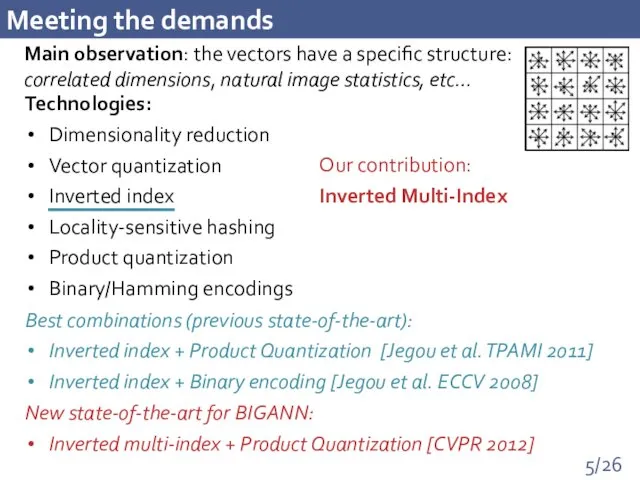
- 5. Meeting the demands Main observation: the vectors have a specific structure: correlated dimensions, natural image statistics,
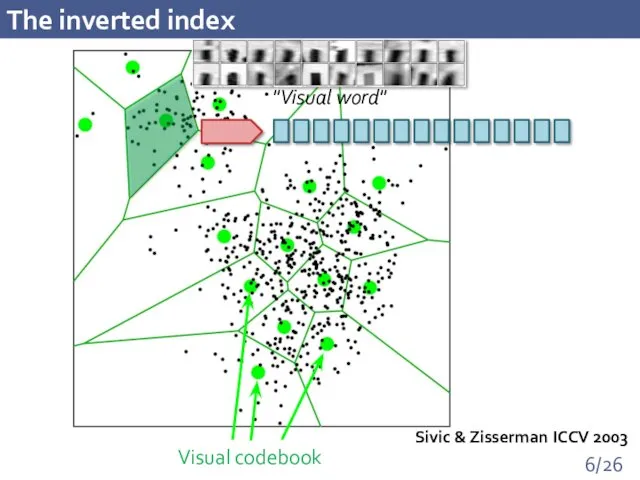
- 6. The inverted index Sivic & Zisserman ICCV 2003
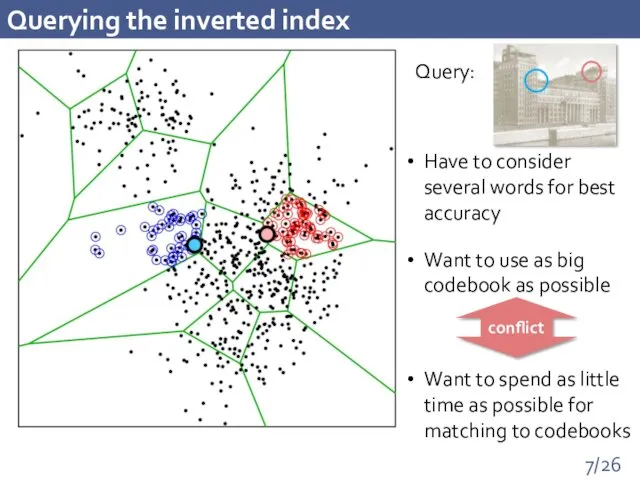
- 7. Querying the inverted index Have to consider several words for best accuracy Want to use as
- 8. Product quantization [Jegou, Douze, Schmid // TPAMI 2011]: Split vector into correlated subvectors use separate small
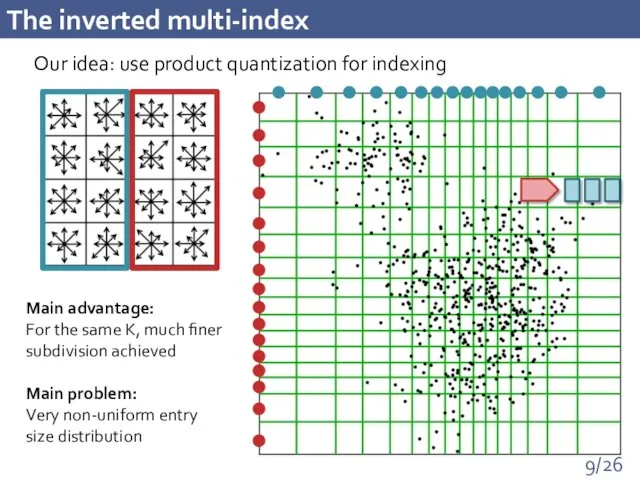
- 9. The inverted multi-index Our idea: use product quantization for indexing Main advantage: For the same K,
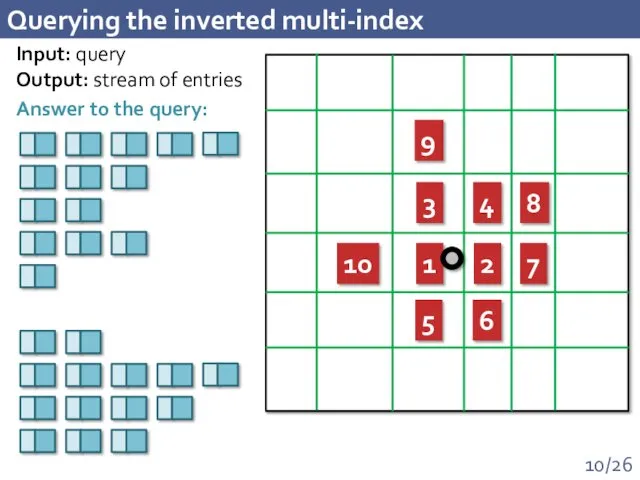
- 10. Querying the inverted multi-index 1 2 3 4 5 6 7 8 9 10 Input: query
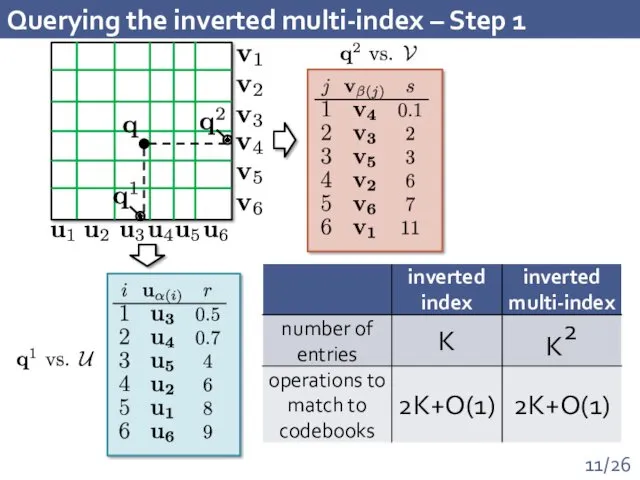
- 11. Querying the inverted multi-index – Step 1
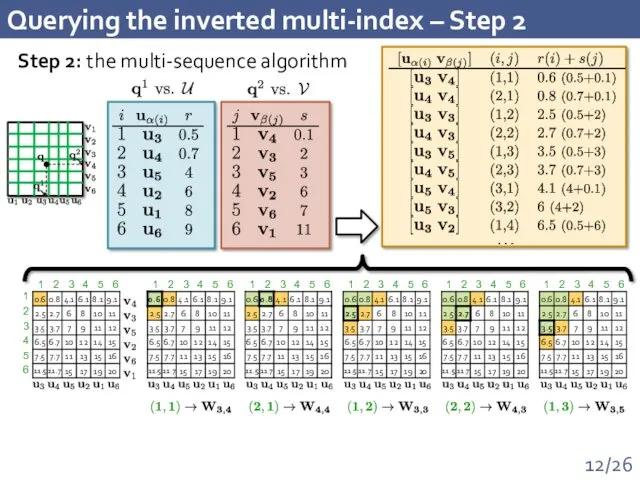
- 12. Querying the inverted multi-index – Step 2 1 2 3 4 5 6 1 2 3
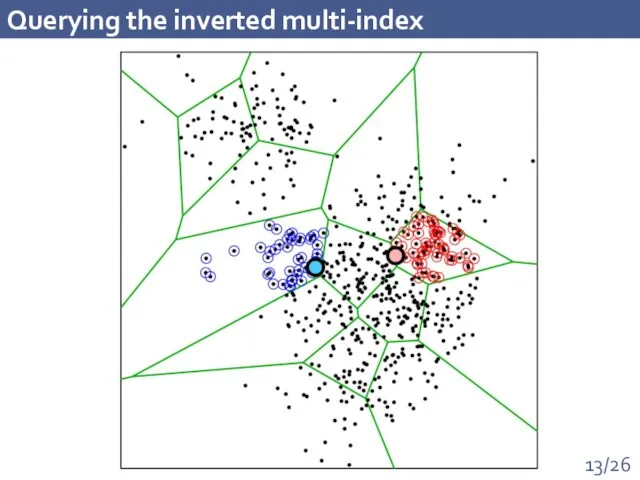
- 13. Querying the inverted multi-index
- 14. Experimental protocol Dataset: 1 billion of SIFT vectors [Jegou et al.] Hold-out set of 10000 queries,
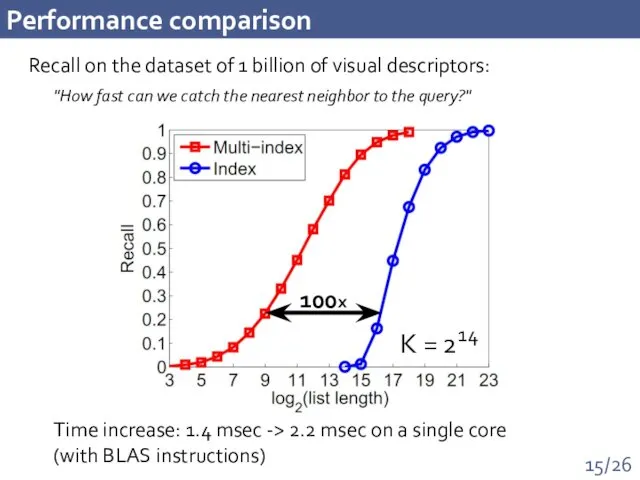
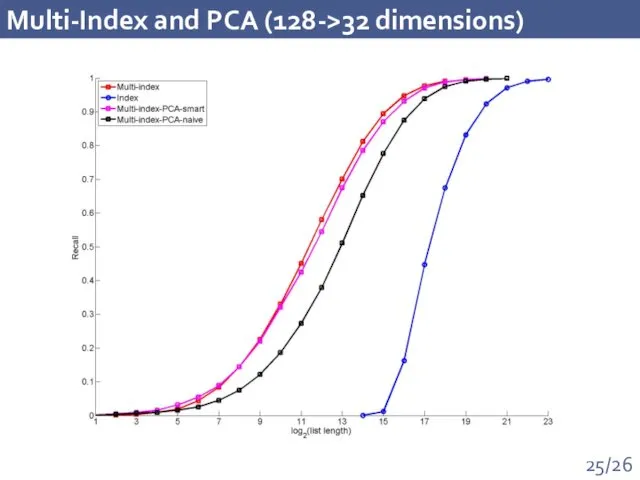
- 15. Performance comparison Recall on the dataset of 1 billion of visual descriptors: 100x Time increase: 1.4
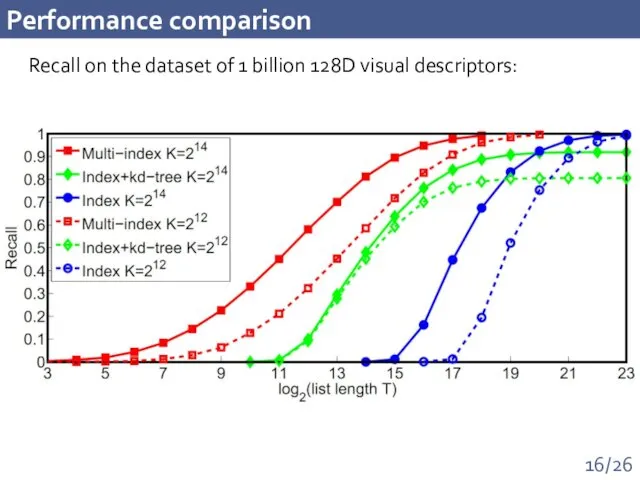
- 16. Performance comparison Recall on the dataset of 1 billion 128D visual descriptors:
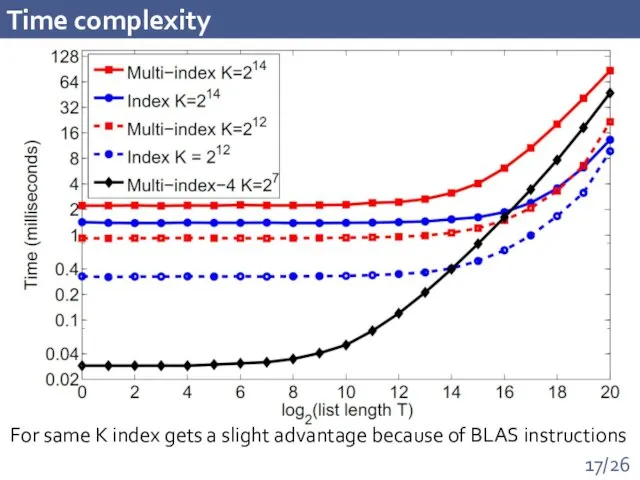
- 17. Time complexity For same K index gets a slight advantage because of BLAS instructions
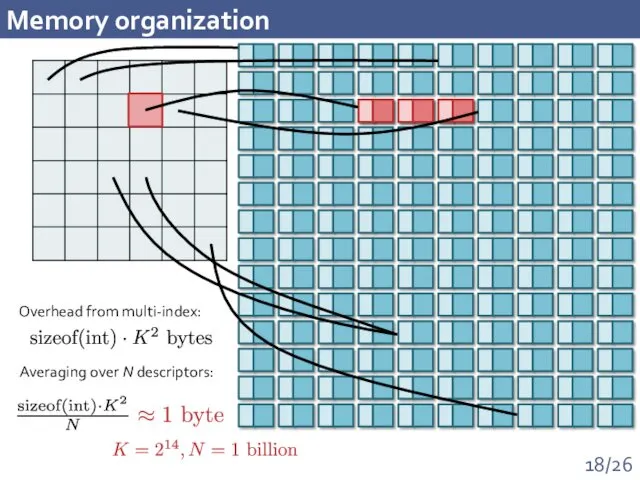
- 18. Memory organization Overhead from multi-index: Averaging over N descriptors:
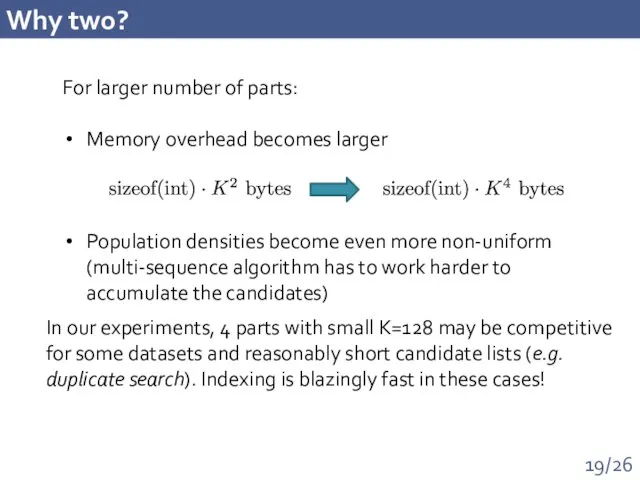
- 19. Why two? For larger number of parts: Memory overhead becomes larger Population densities become even more
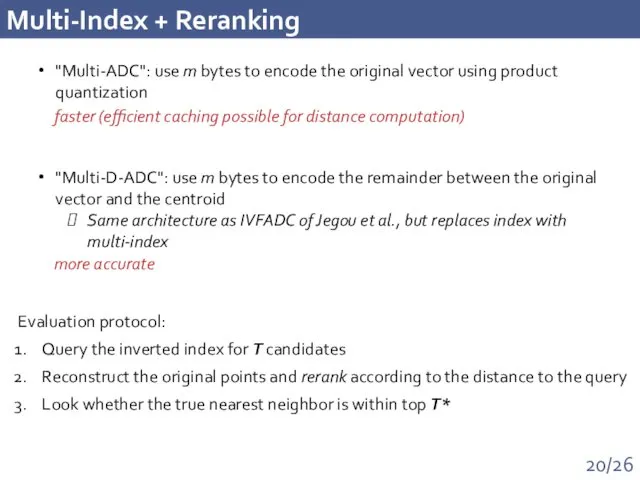
- 20. Multi-Index + Reranking "Multi-ADC": use m bytes to encode the original vector using product quantization "Multi-D-ADC":
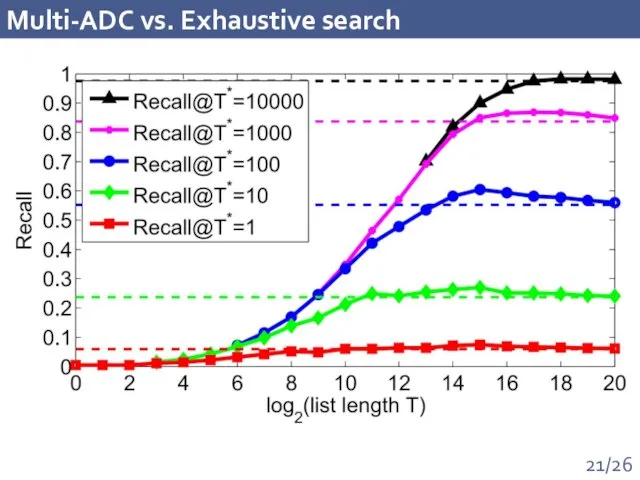
- 21. Multi-ADC vs. Exhaustive search
- 22. Multi-D-ADC vs State-of-the-art State-of-the-art [Jegou et al.] Combining multi-index + reranking:
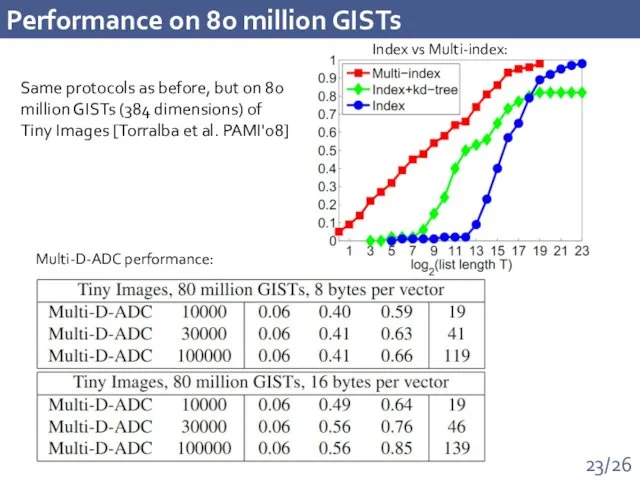
- 23. Performance on 80 million GISTs Multi-D-ADC performance: Index vs Multi-index: Same protocols as before, but on
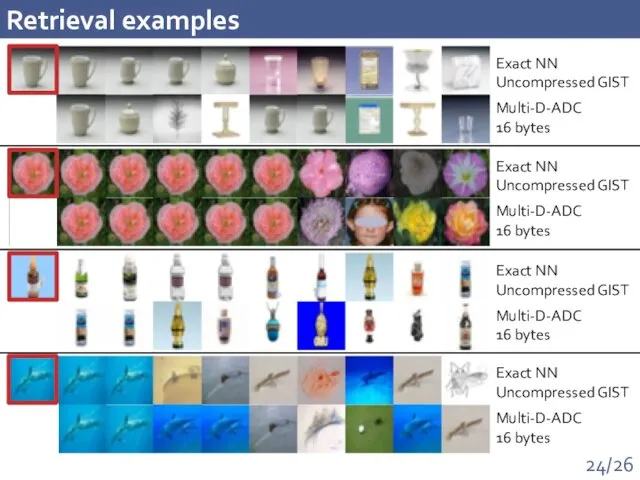
- 24. Retrieval examples Exact NN Uncompressed GIST Multi-D-ADC 16 bytes Exact NN Uncompressed GIST Multi-D-ADC 16 bytes
- 25. Multi-Index and PCA (128->32 dimensions)
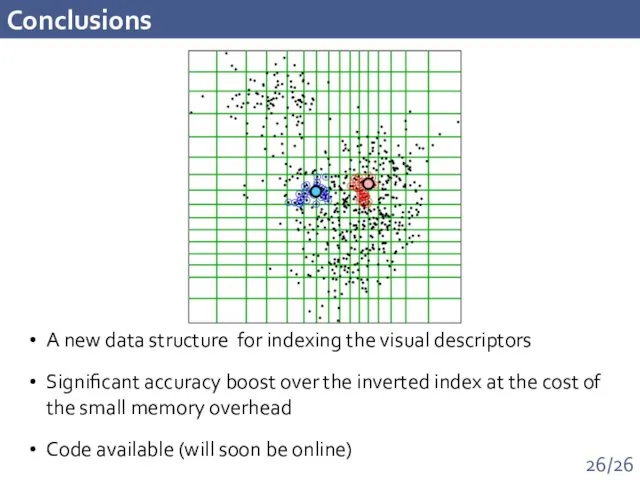
- 26. Conclusions A new data structure for indexing the visual descriptors Significant accuracy boost over the inverted
- 27. Other usage scenarios Large-scale NN search' based approaches: Holistic high dimensional image descriptors: GISTs, VLADs, Fisher
- 29. Скачать презентацию
![Product quantization [Jegou, Douze, Schmid // TPAMI 2011]: Split vector](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/87883/slide-7.jpg)

![Multi-D-ADC vs State-of-the-art State-of-the-art [Jegou et al.] Combining multi-index + reranking:](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/87883/slide-21.jpg)

 Как подготовить данные. Семинар 4. Викторина
Как подготовить данные. Семинар 4. Викторина Концепция и возможности подхода .NET
Концепция и возможности подхода .NET Microsoft Access Мәліметтер қорын басқару жүйесі
Microsoft Access Мәліметтер қорын басқару жүйесі Технология создания и обработки графической информации
Технология создания и обработки графической информации Презентация Времена года. 6 класс
Презентация Времена года. 6 класс Строки Паскаль. Чем плох массив символов?
Строки Паскаль. Чем плох массив символов? Научная информация: поиск, накопление и обработка
Научная информация: поиск, накопление и обработка Обслуживание сети
Обслуживание сети Introduction to Data Capture. Module 6
Introduction to Data Capture. Module 6 Решение логических задач
Решение логических задач Передача информации. Приложение к уроку
Передача информации. Приложение к уроку Добровольцы России. Регистрация организации
Добровольцы России. Регистрация организации Упрощение логических выражений. Решение задач
Упрощение логических выражений. Решение задач ВКР: Совершенствование системы управления персоналом сервисного предприятия
ВКР: Совершенствование системы управления персоналом сервисного предприятия Как снимать интересные сториз
Как снимать интересные сториз Память. Что такое память компьютера
Память. Что такое память компьютера Практические аспекты составления алгоритмов
Практические аспекты составления алгоритмов Теоретические основы информатики
Теоретические основы информатики Информационные технологии в государственном управлении
Информационные технологии в государственном управлении Компьютерлік желі
Компьютерлік желі WEB. Visual Studio Code
WEB. Visual Studio Code Браузеры. Яндекс Браузер, Opera, Firefox
Браузеры. Яндекс Браузер, Opera, Firefox SQL. База данных
SQL. База данных Системы управления базами данных (СУБД) MS Access
Системы управления базами данных (СУБД) MS Access Решение задач с использованием операторов цикла
Решение задач с использованием операторов цикла Лекция 15. Основные механизмы защиты, используемые в системах защиты информации (СЗИ) информационных систем (ИС)
Лекция 15. Основные механизмы защиты, используемые в системах защиты информации (СЗИ) информационных систем (ИС) Графический учебный исполнитель. Разработка урока с презентацией
Графический учебный исполнитель. Разработка урока с презентацией Программирование на языке Java. Тема 23. Рекурсия
Программирование на языке Java. Тема 23. Рекурсия