Содержание
- 2. In vivo (in the cell): - RNA-encoded protein chain is synthesized at a ribosome. - Biosynthesis
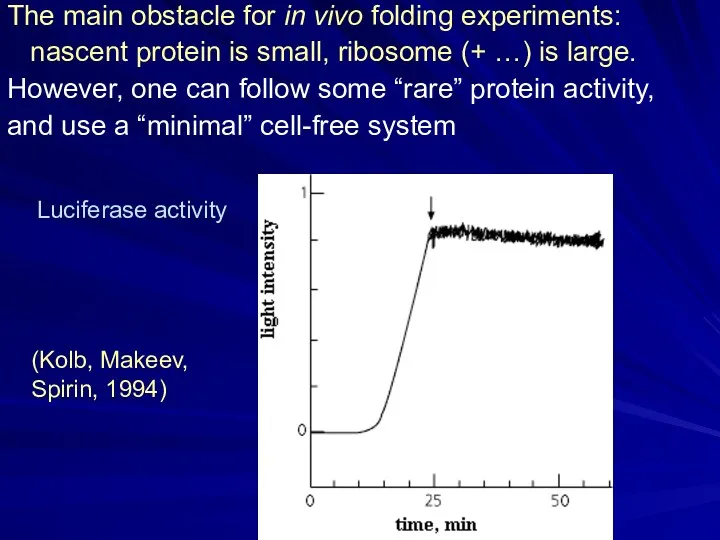
- 3. The main obstacle for in vivo folding experiments: nascent protein is small, ribosome (+ …) is
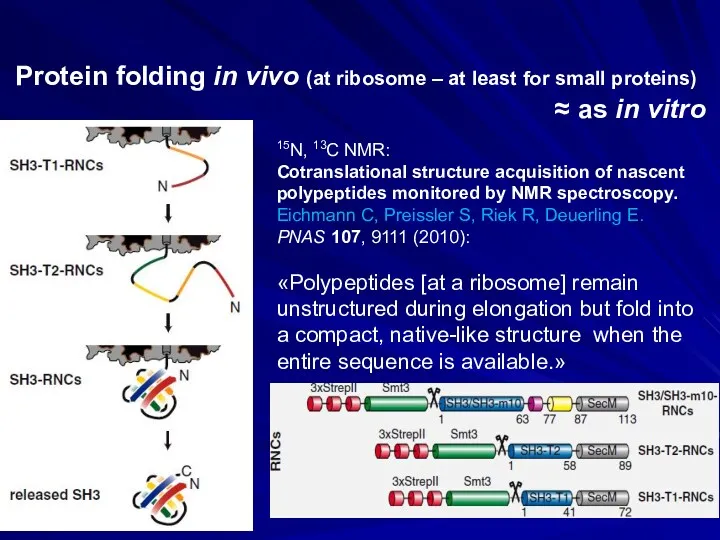
- 4. 15N, 13C NMR: Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Eichmann C, Preissler
- 5. 15N, 13C NMR: Monitoring cotranslational protein folding in mammalian cells at codon resolution. Han Y., David
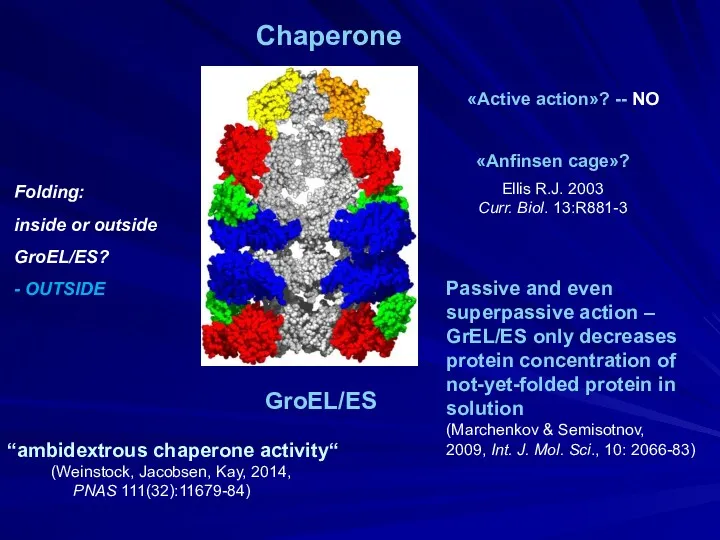
- 6. Folding: inside or outside GroEL/ES? - OUTSIDE «Anfinsen cage»? Ellis R.J. 2003 Curr. Biol. 13:R881-3 Passive
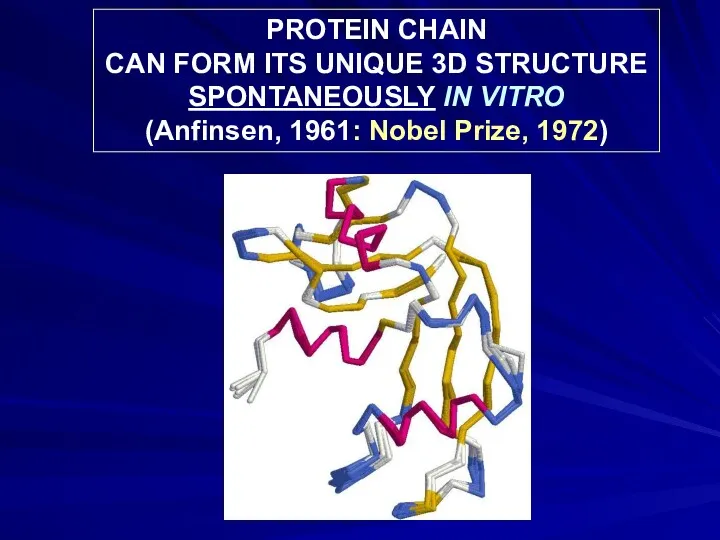
- 7. PROTEIN CHAIN CAN FORM ITS UNIQUE 3D STRUCTURE SPONTANEOUSLY IN VITRO (Anfinsen, 1961: Nobel Prize, 1972)
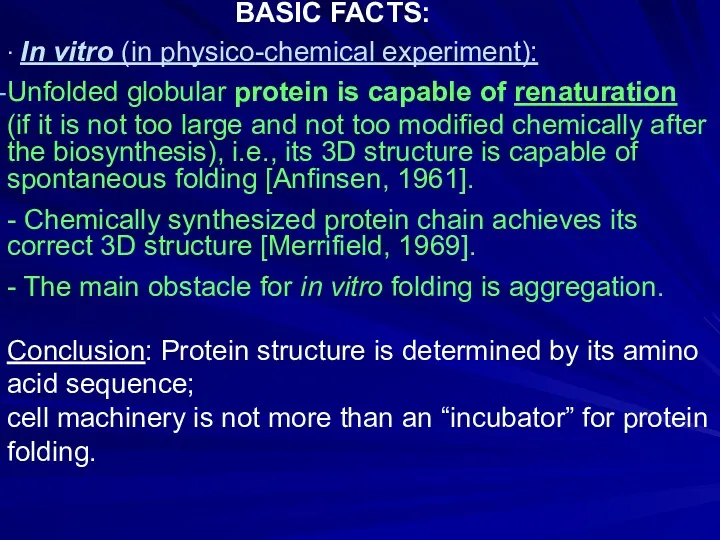
- 8. ∙ In vitro (in physico-chemical experiment): Unfolded globular protein is capable of renaturation (if it is
- 9. Robert Bruce Merrifield (1921 – 2006) Nobel Prize 1988 Christian Boehmer Anfinsen, Jr. (1916 –1995) Nobel
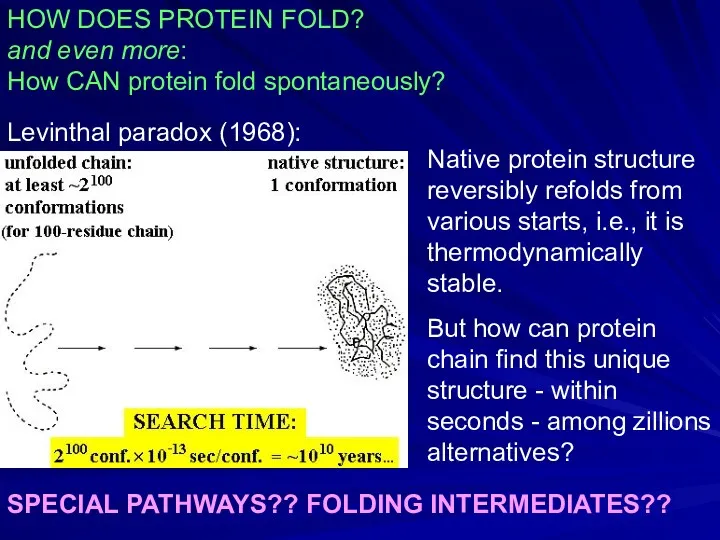
- 10. HOW DOES PROTEIN FOLD? and even more: How CAN protein fold spontaneously? Levinthal paradox (1968): SPECIAL
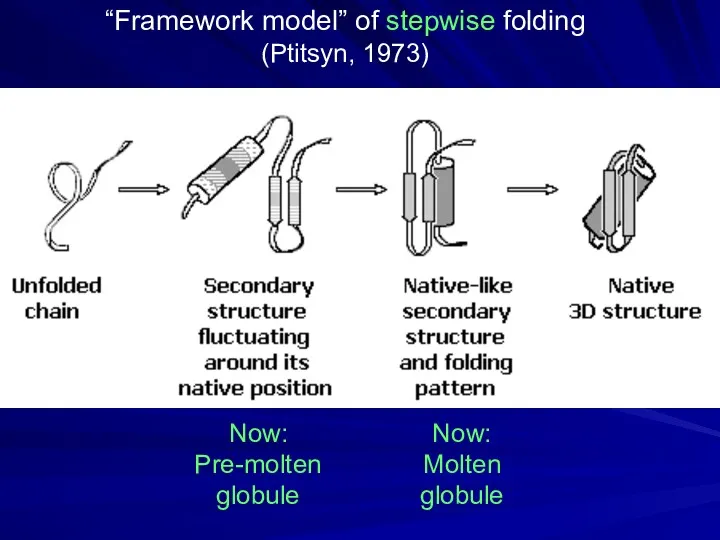
- 11. “Framework model” of stepwise folding (Ptitsyn, 1973) Now: Pre-molten globule Now: Molten globule
- 12. Oleg Borisovich Ptitsyn (1929-99)
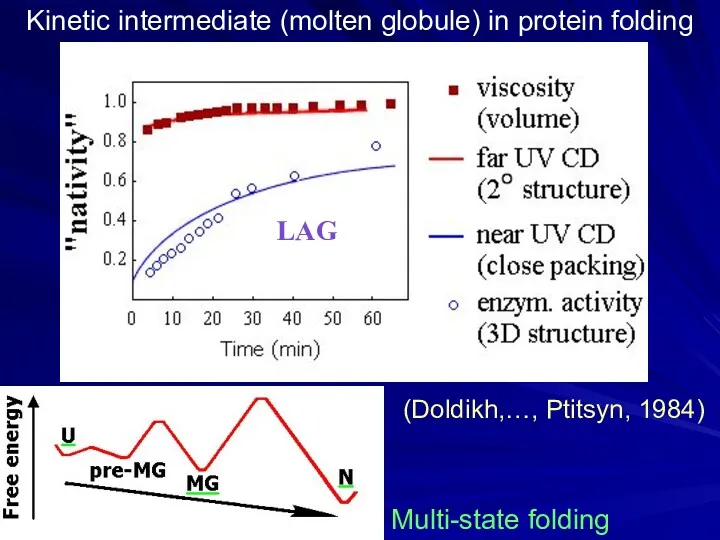
- 13. Kinetic intermediate (molten globule) in protein folding (Doldikh,…, Ptitsyn, 1984) Multi-state folding LAG
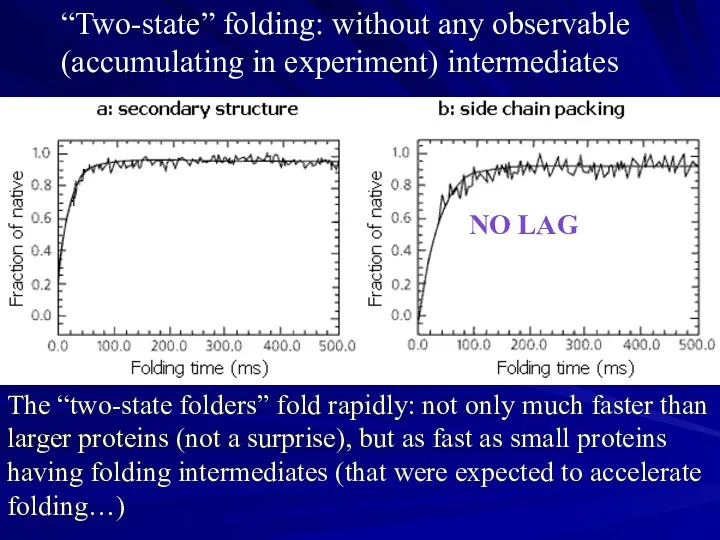
- 14. Found: metastable (“accumulating”, “directly observable”) folding intermediates. The idea was: intermediates will help to trace the
- 15. “Two-state” folding: without any observable (accumulating in experiment) intermediates The “two-state folders” fold rapidly: not only
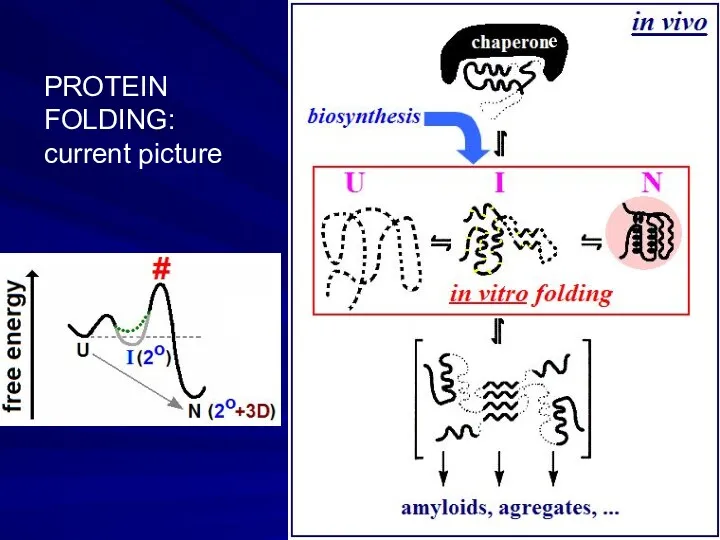
- 16. e PROTEIN FOLDING: current picture
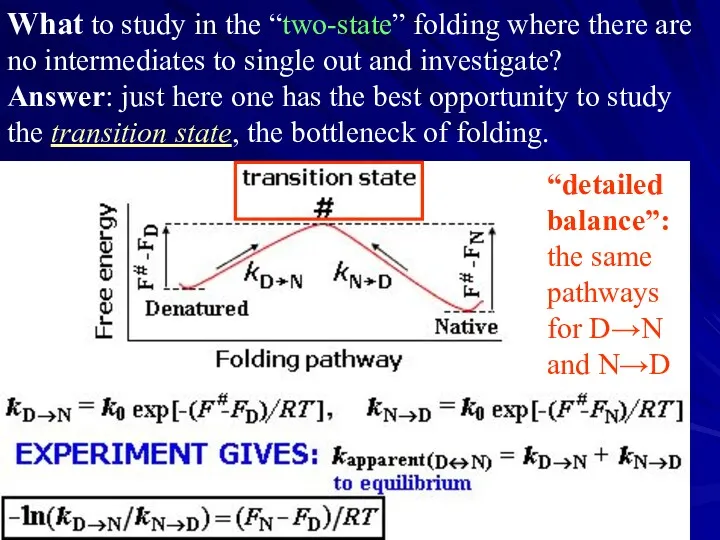
- 17. What to study in the “two-state” folding where there are no intermediates to single out and
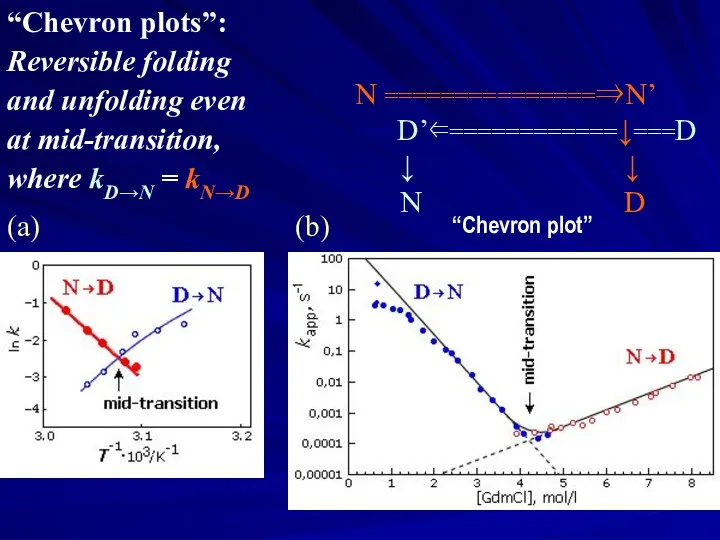
- 18. “Chevron plots”: Reversible folding and unfolding even at mid-transition, where kD→N = kN→D (a) (b) N
- 19. Sir Alan Roy Fersht, 1943 Protein engineering Folding nucleus
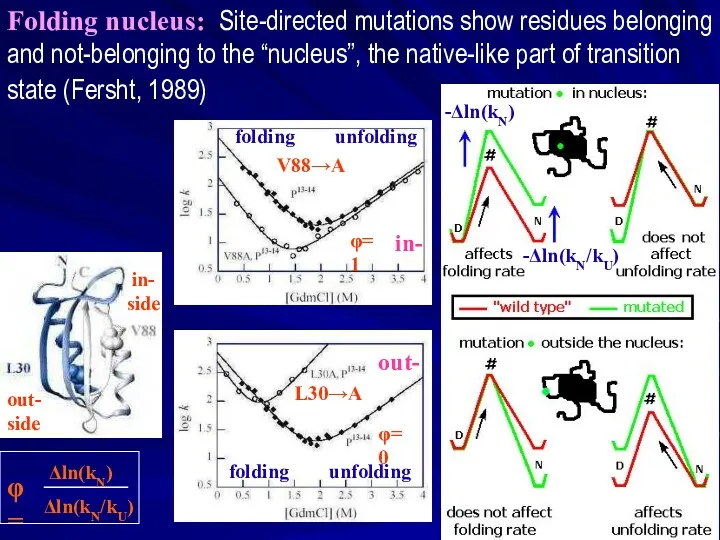
- 20. Folding nucleus: Site-directed mutations show residues belonging and not-belonging to the “nucleus”, the native-like part of
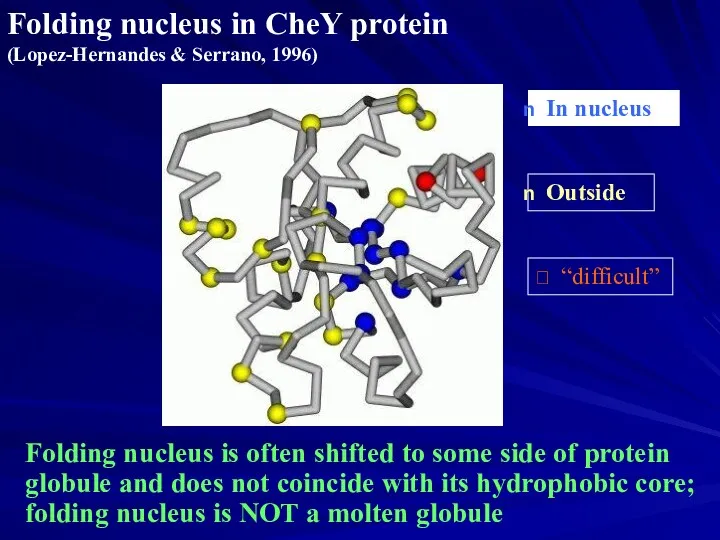
- 21. Folding nucleus in CheY protein (Lopez-Hernandes & Serrano, 1996) In nucleus Outside “difficult” Folding nucleus
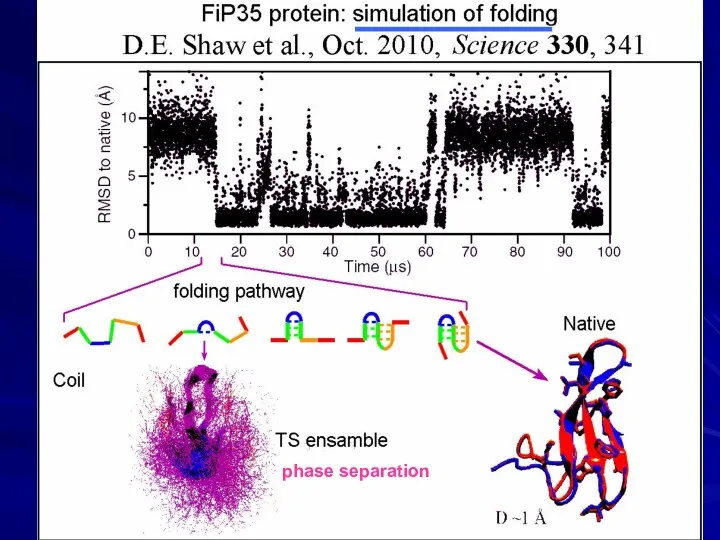
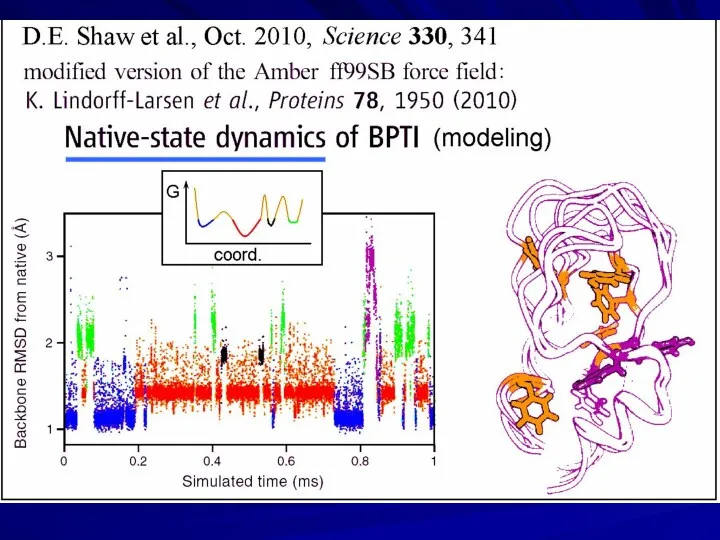
- 22. David E. Shaw “D. E. Shaw Research” US$ 3.5 billion Supercomputer “Anton” “Hot point” in protein
- 23. phase separation
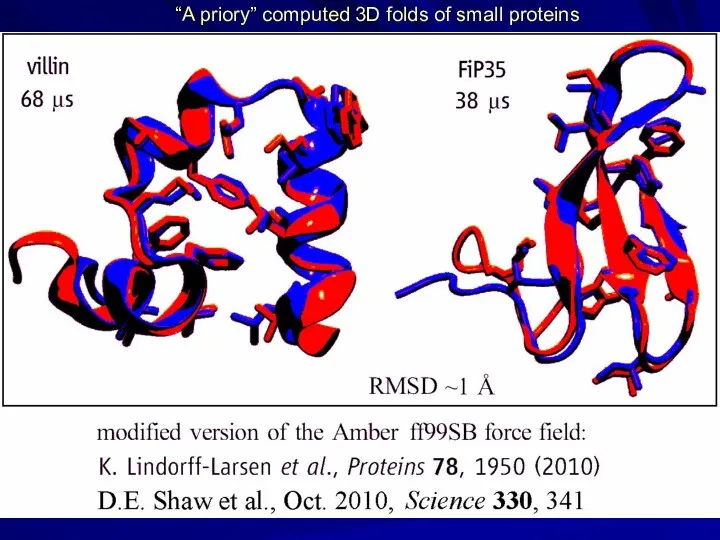
- 24. “A priory” computed 3D folds of small proteins
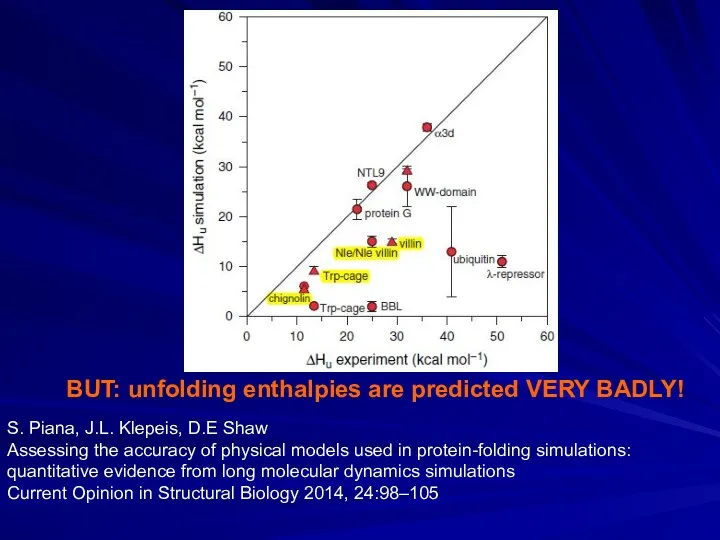
- 25. BUT: unfolding enthalpies are predicted VERY BADLY! S. Piana, J.L. Klepeis, D.E Shaw Assessing the accuracy
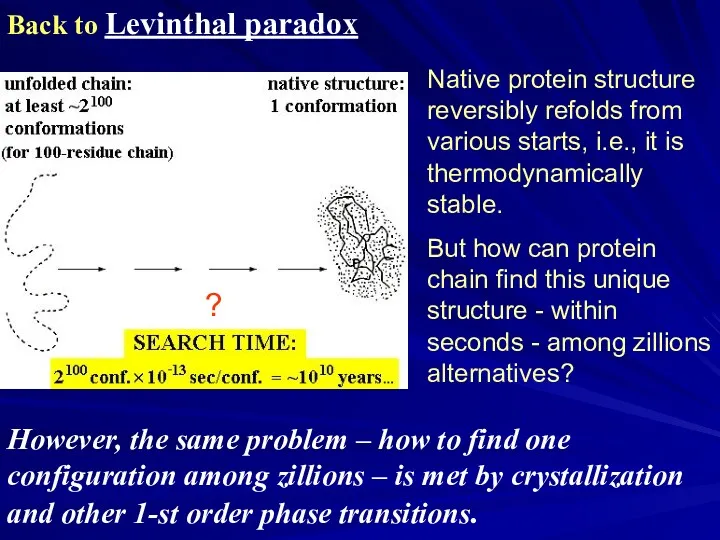
- 26. Back to Levinthal paradox Native protein structure reversibly refolds from various starts, i.e., it is thermodynamically
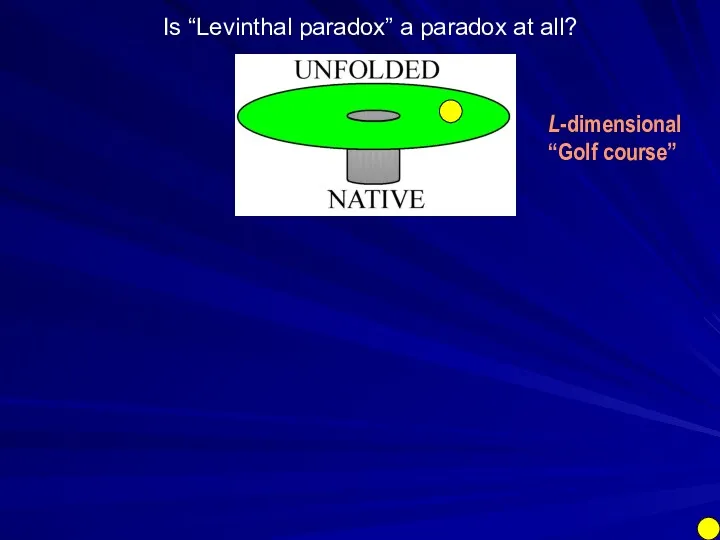
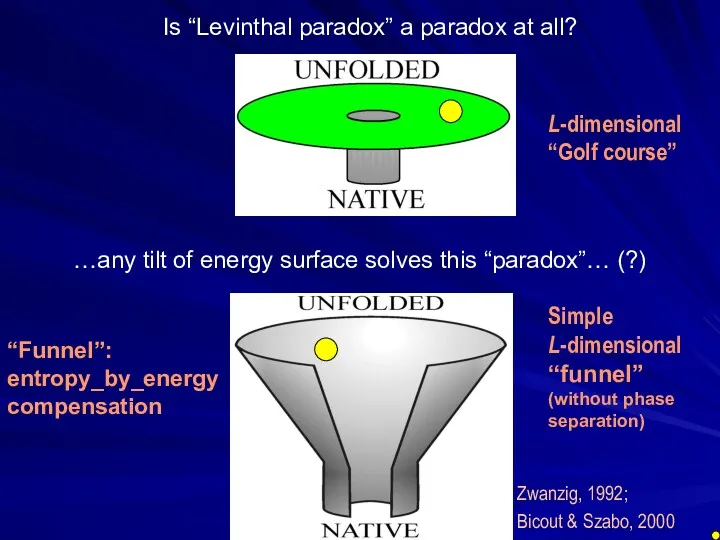
- 27. Is “Levinthal paradox” a paradox at all? L-dimensional “Golf course”
- 28. Zwanzig, 1992; Bicout & Szabo, 2000 Is “Levinthal paradox” a paradox at all? …any tilt of
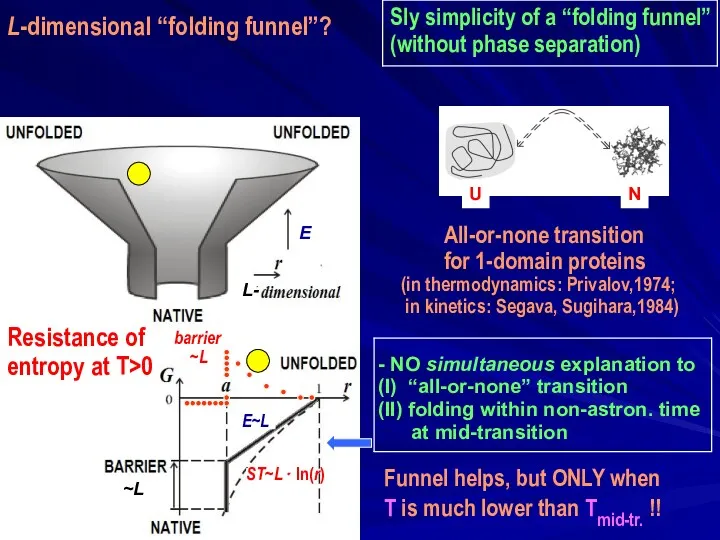
- 29. Sly simplicity of a “folding funnel” (without phase separation) - NO simultaneous explanation to (I) “all-or-none”
- 30. Phillips (1965) hypothesis: folding nucleus is formed by the N-end of the nascent protein chain, and
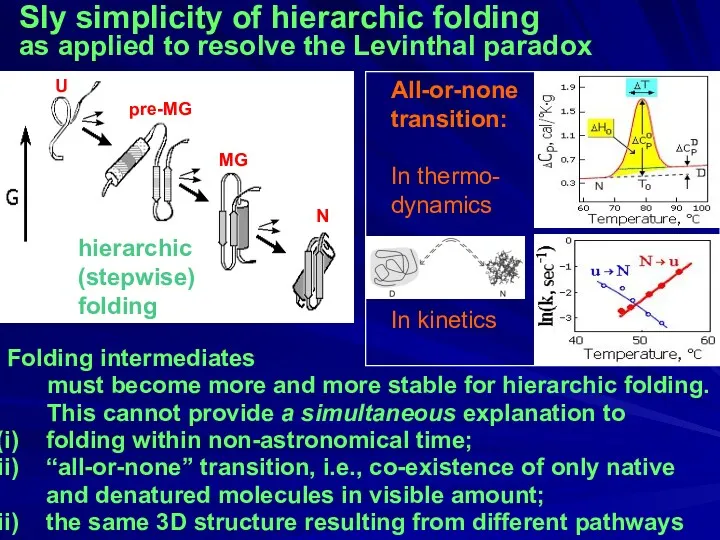
- 31. Sly simplicity of hierarchic folding as applied to resolve the Levinthal paradox Folding intermediates must become
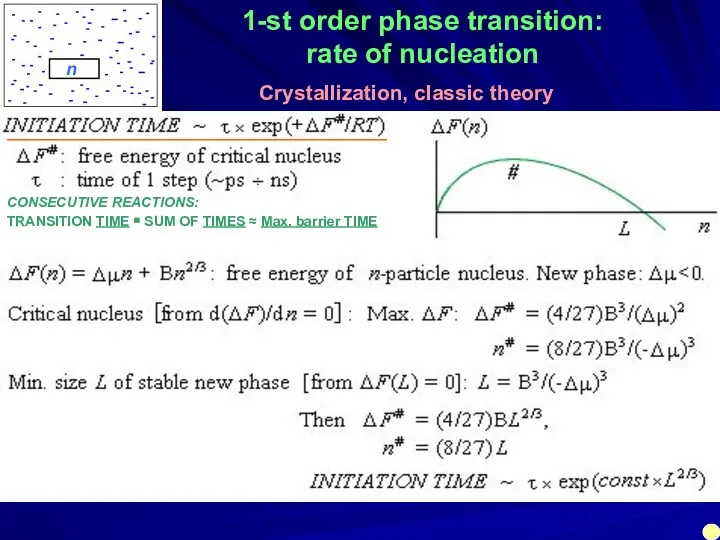
- 32. 1-st order phase transition: rate of nucleation Crystallization, classic theory n ______________________________________ CONSECUTIVE REACTIONS: TRANSITION TIME
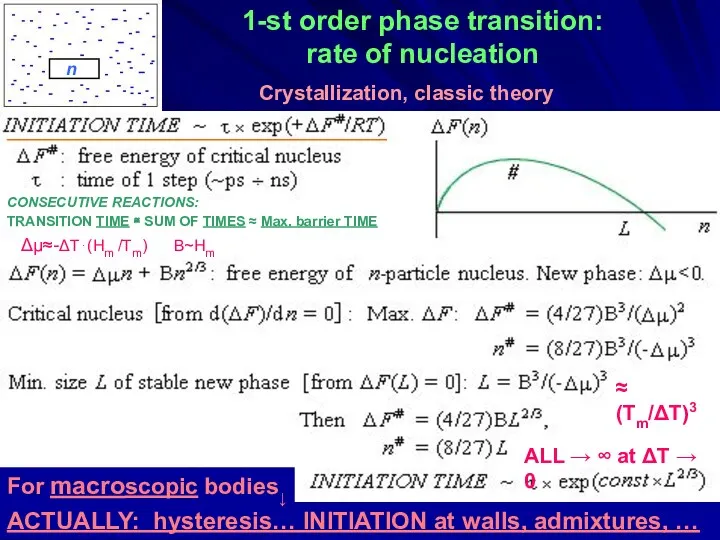
- 33. 1-st order phase transition: rate of nucleation Δμ≈-ΔT⋅(Hm /Tm) B~Hm ≈ (Tm/ΔT)3 ALL → ∞ at
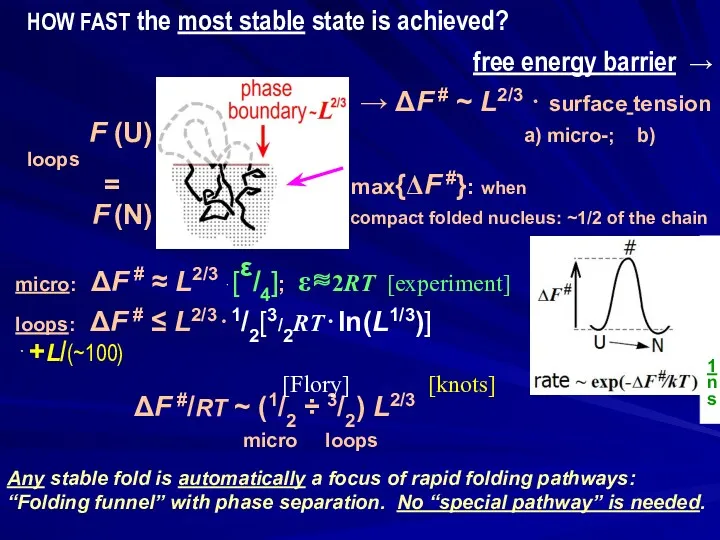
- 34. Let us consider sequential folding (or unfolding) of a chain that has a large energy gap
- 35. L 1 ns ΔF #/RT ~ (1/2 ÷ 3/2) L2/3 micro loops Any stable fold is
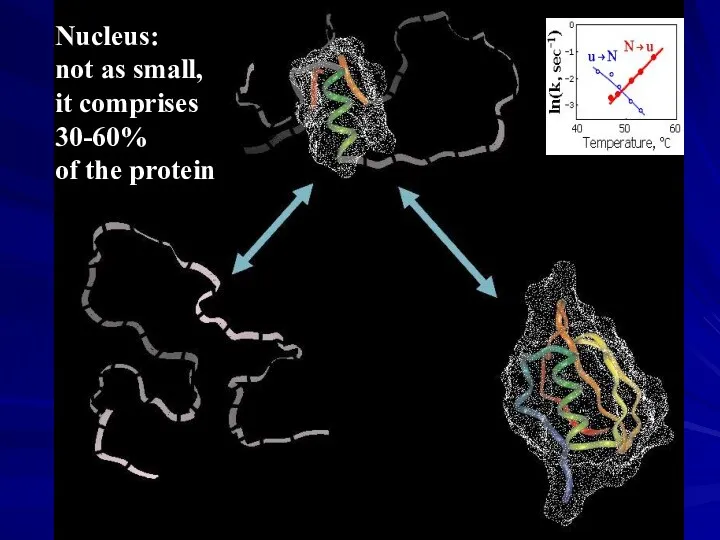
- 36. Nucleus: not as small, it comprises 30-60% of the protein
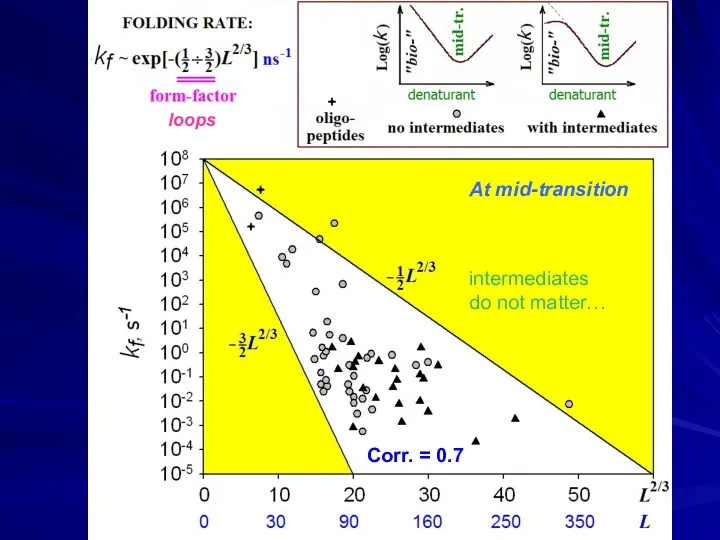
- 37. ↓ ↓ Corr. = 0.7 loops At mid-transition intermediates do not matter…
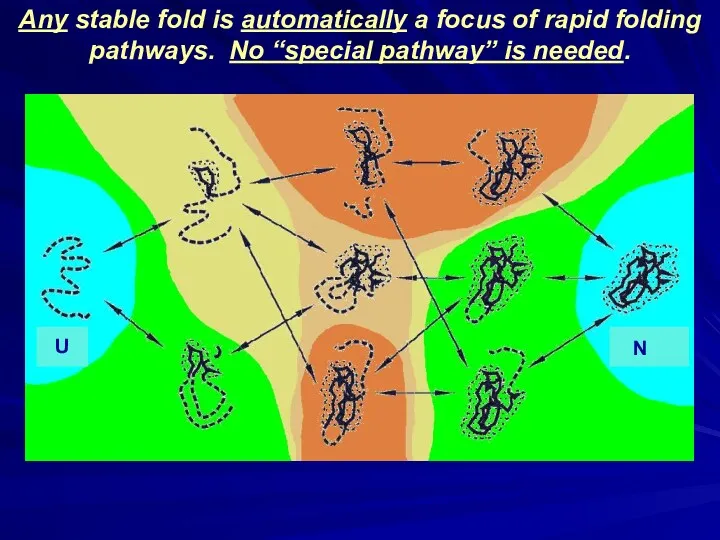
- 38. ↓ ↓ ↓ ΔFN ↓ ↓ ΔFN ↓ Any stable fold is automatically a focus of
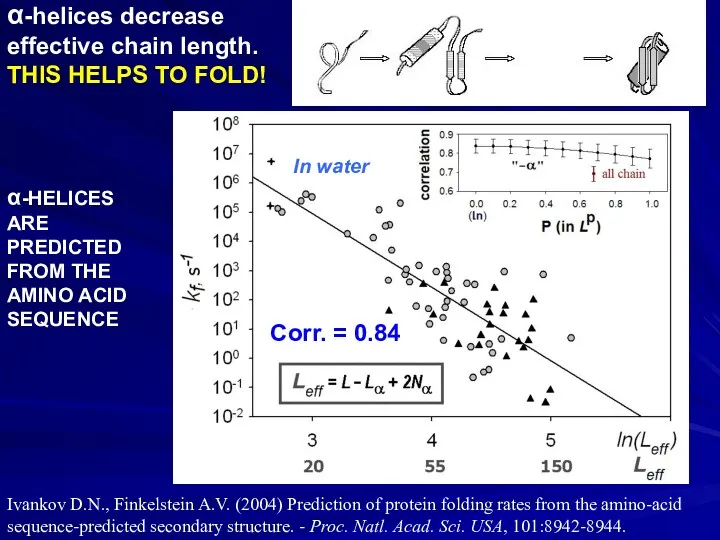
- 39. α-helices decrease effective chain length. THIS HELPS TO FOLD! Corr. = 0.84 α-HELICES ARE PREDICTED FROM
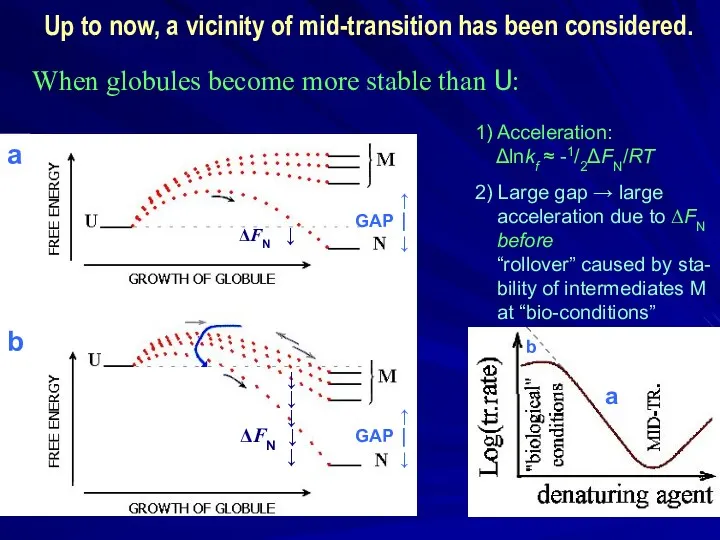
- 40. When globules become more stable than U: a b a b ↑ GAP ⏐ ↓ 1)
- 41. Finkelstein, Badretdinov; Folding & Design, 1997, 1998]. Finkelstein; Les Houches, Session 77, 2003] Garbuzynskiy, Ivankov, Bogatyreva,
- 42. Finkelstein, Badretdinov; Folding & Design, 1997, 1998]. Finkelstein; Les Houches, Session 77, 2003] Garbuzynskiy, Ivankov, Bogatyreva,
- 45. Скачать презентацию









![Finkelstein, Badretdinov; Folding & Design, 1997, 1998]. Finkelstein; Les Houches,](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/249784/slide-40.jpg)
![Finkelstein, Badretdinov; Folding & Design, 1997, 1998]. Finkelstein; Les Houches,](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/249784/slide-41.jpg)
 Гидравлика
Гидравлика урок-игра по физике по теме Плотность
урок-игра по физике по теме Плотность Анализ технического паспорта асинхронного электродвигателя
Анализ технического паспорта асинхронного электродвигателя Мегаконструкції. Найбільший в світі екскаватор
Мегаконструкції. Найбільший в світі екскаватор Современные автомобили и двигатели
Современные автомобили и двигатели Внедрение ФГОС общего образования второго поколения по физике
Внедрение ФГОС общего образования второго поколения по физике Технология мини-исследования на уроках физики
Технология мини-исследования на уроках физики спектры
спектры Bbs wheels range 1999 ilustrated catalog
Bbs wheels range 1999 ilustrated catalog Законам механики подчиняются движения всех окружающих нас тел
Законам механики подчиняются движения всех окружающих нас тел Конкурс учителей Есть идея!
Конкурс учителей Есть идея! Олимпийские игры и физика
Олимпийские игры и физика Памятка по неисправностям автосцепного устройства при техническом обслуживании
Памятка по неисправностям автосцепного устройства при техническом обслуживании Урок физики в 10 классе Газовые законы
Урок физики в 10 классе Газовые законы Звуковые волны
Звуковые волны Элементарные частицы. Микромир
Элементарные частицы. Микромир Идеальный газ. Основное уравнение МКТ идеального газа
Идеальный газ. Основное уравнение МКТ идеального газа Нормирование точности поверхностей деталей машин по взаимному расположению, обозначения их на чертежах
Нормирование точности поверхностей деталей машин по взаимному расположению, обозначения их на чертежах презентация по теме Сила трения.
презентация по теме Сила трения. Стискання газів. Рівняння Менделєєва-Клапейрона
Стискання газів. Рівняння Менделєєва-Клапейрона Axles and shafts
Axles and shafts дополнение к уроку физики Испарение
дополнение к уроку физики Испарение Изобретени радио А.С. Поповым
Изобретени радио А.С. Поповым Гармонические колебания и их характеристики
Гармонические колебания и их характеристики Цепные передачи
Цепные передачи Mechanism
Mechanism Classical angular momentum and magnetic dipole moment
Classical angular momentum and magnetic dipole moment Кинематика вращательного движения
Кинематика вращательного движения