Слайд 2

From the very beginning
...AACCCGTACGTTTTGCAAACGACCGT...
Слайд 3

From the very beginning
Sequencing
...AACCCGTACGTTTTGCAAACGACCGT...
AACCCGTACGT
CGTACGTTTTG
AACGACCG
GTTTTGCAAACG
GTACGTTTTGCA
Слайд 4
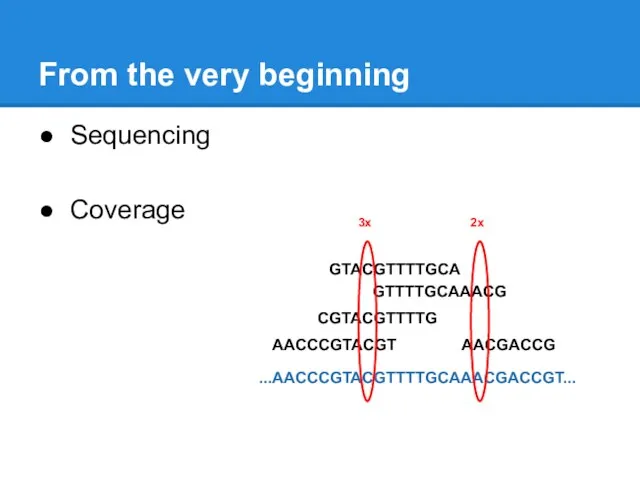
From the very beginning
Sequencing
Coverage
...AACCCGTACGTTTTGCAAACGACCGT...
AACCCGTACGT
CGTACGTTTTG
AACGACCG
GTTTTGCAAACG
GTACGTTTTGCA
3x
2x
Слайд 5

From the very beginning
Sequencing
Coverage
Errors
Mismatches
...AACCCGTACGTTTTGCAAACGACCGT...
AACCCGTTCGT
CGTACGTTTTC
AACGACCG
GTTTTGCAAACG
GTACGTTTTGCA
Слайд 6

From the very beginning
Sequencing
Coverage
Errors
Mismatches
Indels
...AACCCGTACGTTTTGCAAACGACCGT...
AACCCGTTCGT
CGTACGTTTTTC
AACGACCG
GTTTTGCAAACG
GTA_GTTTTGCA
Слайд 7

Early days
Sanger sequencing
Long reads (~900 bp)
Low coverage (< 10x)
Extreme cost
Human genome
project
3 Gbp
3 billion USD
10 years
Слайд 8

NGS
Shorter reads (25-400bp)
High coverage (50-1000x)
Huge amount of data
Low cost
More applications
Required
completely new algorithms
Слайд 9
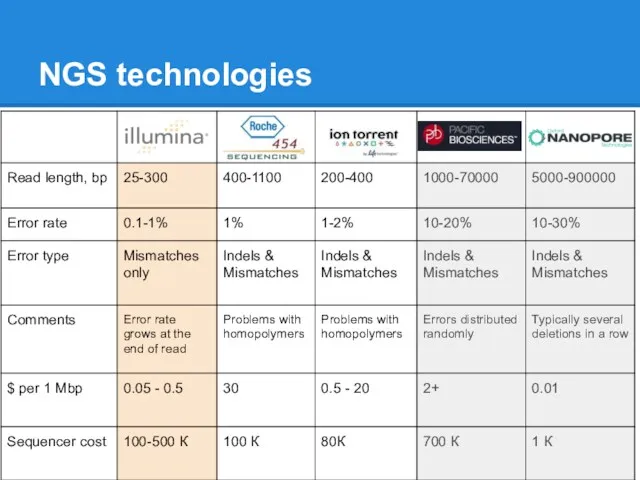
Слайд 10
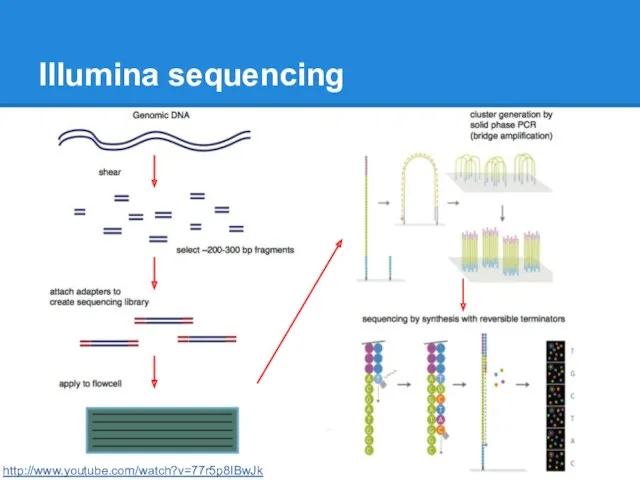
Illumina sequencing
http://www.youtube.com/watch?v=77r5p8IBwJk
Слайд 11
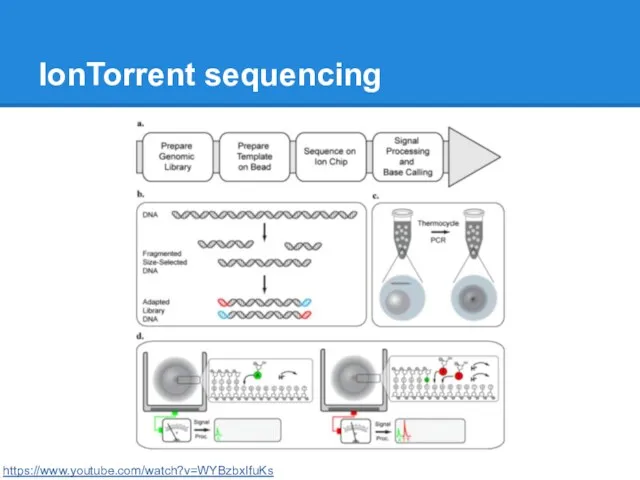
IonTorrent sequencing
https://www.youtube.com/watch?v=WYBzbxIfuKs
Слайд 12
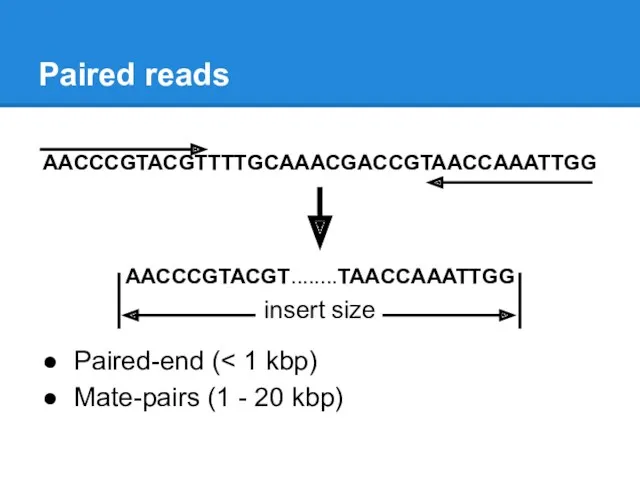
Paired reads
AACCCGTACGTTTTGCAAACGACCGTAACCAAATTGG
AACCCGTACGT........TAACCAAATTGG
insert size
Paired-end (< 1 kbp)
Mate-pairs (1 - 20 kbp)
Слайд 13
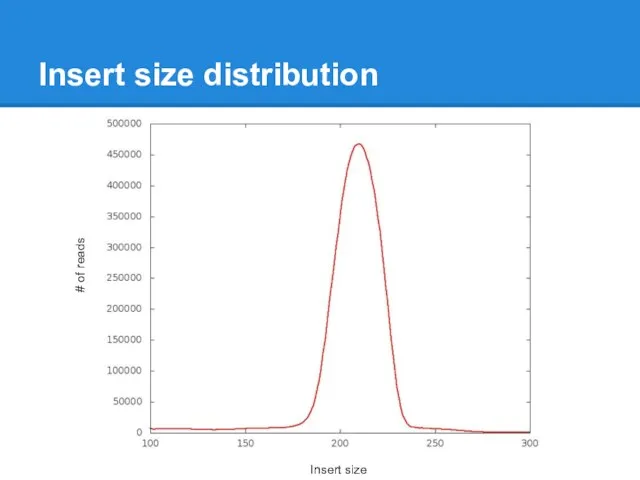
Insert size distribution
Insert size
# of reads
Слайд 14

FASTA/FASTQ
FASTA
>EAS20_8_6_1_9_1972/1
ACCACCATTACCACCACCATCACCATTACCACAGGTAACGGTGCGGGCTGACGC
>EAS20_8_6_1_163_1521/1
GCAGAAAACGTTCTGCATTTGCCACTGATGTACCGCCGAACTTCAACACTCGCA
FASTQ
@EAS20_8_6_1_1477_92/1
ACCGTTACCTGTGGTAATGGTGATGGTGGTGGTAATGGTGGTGCTAATGCGTTT
+EAS20_8_6_1_1477_92/1
HHGHFHHHHHHHHHGFFHHHBG?GGC8DD9GF??=FFBCGBAF>FGCFHGHGGG
Phred quality
Q = [ - 10 log10 p / (1 -
p) ]
Слайд 15

seqtk utility
Subsampling
sample
Converting between interleaved/paired files
mergepe, seq -1/-2
fastq->fasta
seq -A
Quality trimming
Shifting the quality
Modifying
names
etc...
Слайд 16

Слайд 17

FastQC
Easy and lightweight quality control for sequencing data
Does not require reference
genome
Слайд 18
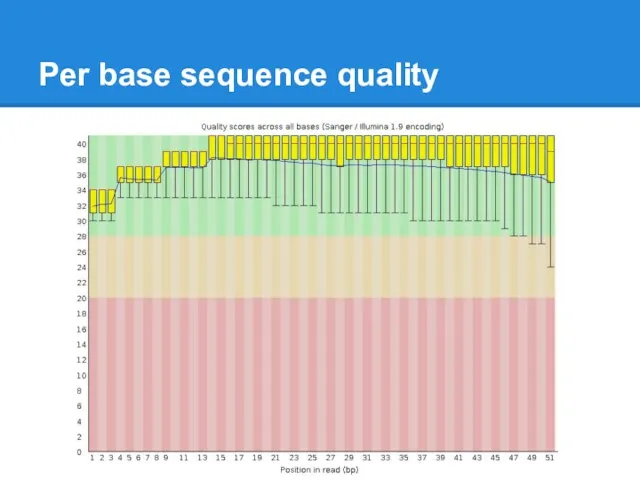
Per base sequence quality
Слайд 19
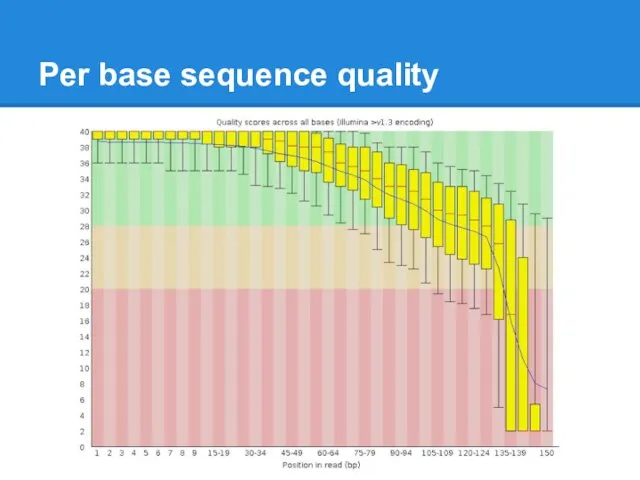
Per base sequence quality
Слайд 20
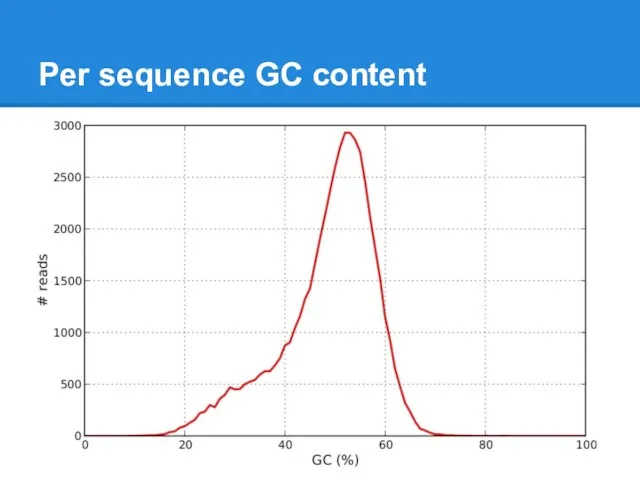
Слайд 21
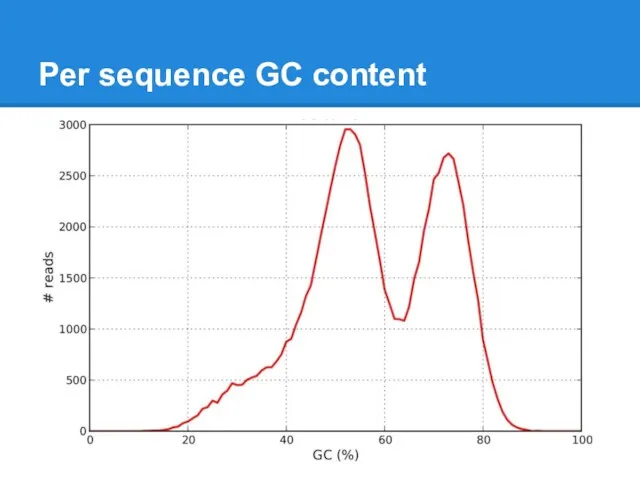
Слайд 22
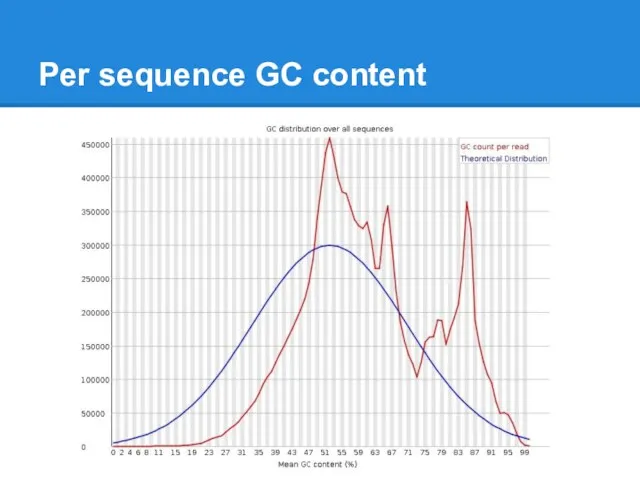
Слайд 23
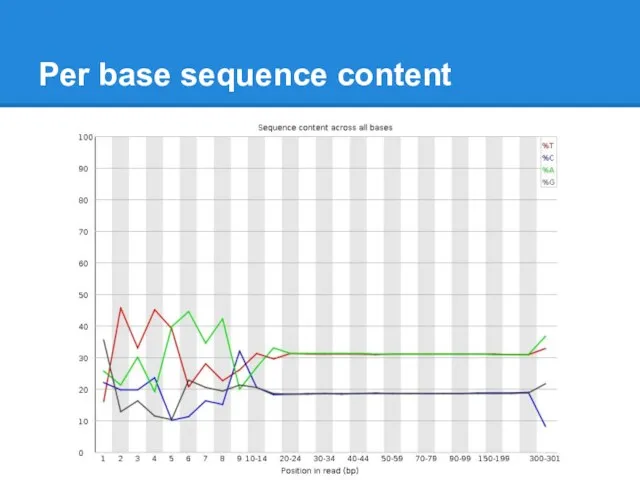
Per base sequence content
Слайд 24
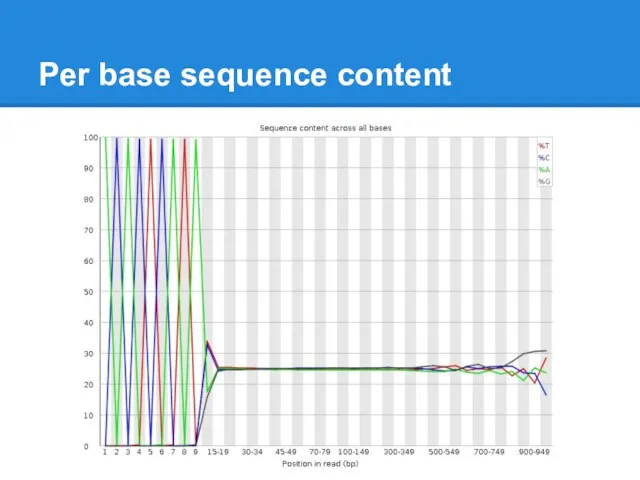
Per base sequence content
Слайд 25

FastQC
fastqc -h
mkdir
Слайд 26
Слайд 27
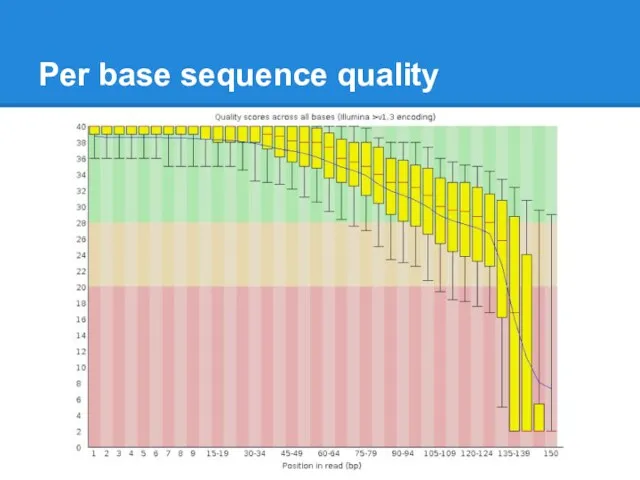
Per base sequence quality
Слайд 28

quality or N bases (below quality 3) (LEADING:3)
Remove trailing low quality or N bases (below quality 3) (TRAILING:3)
Слайд 29

Trimmomatic
Scan the read with a 4-base wide sliding window, cutting when
the average quality per base drops below 15 (SLIDINGWINDOW:4:15)
Drop reads below the 36 bases long (MINLEN:36)
Слайд 30

OPTIONS
ILLUMINACLIP:
ILLUMINACLIP:TruSeq3-PE.fa
Слайд 31

Adapter trimming
ILLUMINACLIP:::threshold>:
ILLUMINACLIP:NexteraPE-PE.fa:2:10:30






























 Лекции 1-2. Введение в моделирование данных, базы данных и SQL
Лекции 1-2. Введение в моделирование данных, базы данных и SQL Тема 2. Каскадные таблицы стилей CSS
Тема 2. Каскадные таблицы стилей CSS Кодирование данных
Кодирование данных Components of computer
Components of computer Понятие информационной системы (ИС). Классификация ИС
Понятие информационной системы (ИС). Классификация ИС AJAX – SPA приложение
AJAX – SPA приложение Konsep Dasar Algoritma
Konsep Dasar Algoritma Продвижение в Instagram
Продвижение в Instagram Графический пакет Adobe Photoshop
Графический пакет Adobe Photoshop Основные компоненты компьютера и их функции
Основные компоненты компьютера и их функции Review or research in software defect reporting
Review or research in software defect reporting Протоколы IPSec
Протоколы IPSec Визитная карточка Свердловской городской библиотеки ЩРБИЦ Щелковского муниципального района
Визитная карточка Свердловской городской библиотеки ЩРБИЦ Щелковского муниципального района Облачные хранилища данных
Облачные хранилища данных Билл Гейтс
Билл Гейтс Аддитивные технологии
Аддитивные технологии Представления о технических и программных средствах телекоммуникационных технологий. Лекция 33. Тема 5.1
Представления о технических и программных средствах телекоммуникационных технологий. Лекция 33. Тема 5.1 Adobe Premier Pro CC. Импорт материалов. Панели мониторинга
Adobe Premier Pro CC. Импорт материалов. Панели мониторинга Среда программирования QBASIK
Среда программирования QBASIK Диалоги и диалоговые окна. Диалоговые окна Windows
Диалоги и диалоговые окна. Диалоговые окна Windows Информационная безопасность
Информационная безопасность Информационные хранилища
Информационные хранилища Представление знаний в экспертных системах
Представление знаний в экспертных системах 01. Введение. Лексика языка Java
01. Введение. Лексика языка Java Построение диаграмм в электронных таблицах
Построение диаграмм в электронных таблицах PHP-де файлдармен барлық
PHP-де файлдармен барлық Дешифрирование космических снимков с использованием ПО ERDAS Imagine
Дешифрирование космических снимков с использованием ПО ERDAS Imagine Введение в разработку мобильных приложений Введение в разработку приложений для смартфонов на ОС Android
Введение в разработку мобильных приложений Введение в разработку приложений для смартфонов на ОС Android