Методы исследования взаимодействий с участием белков (Co-IP, equilibrium microdialysis, ITC, MST, SPR, BLI, QСM) презентация
Содержание
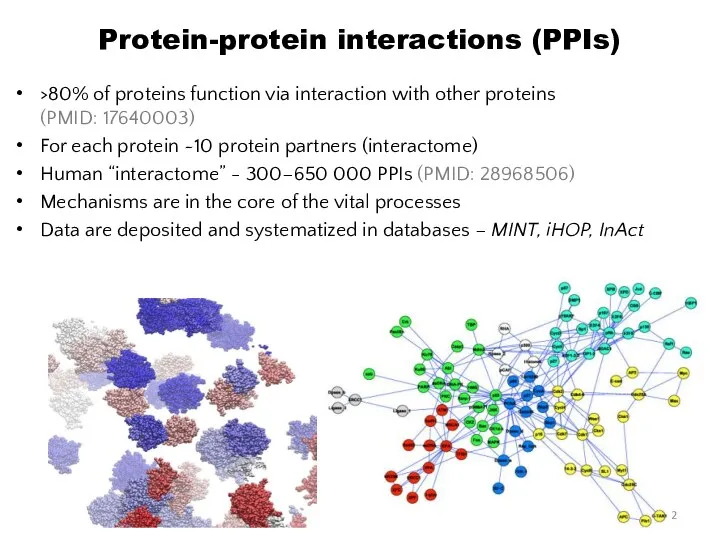
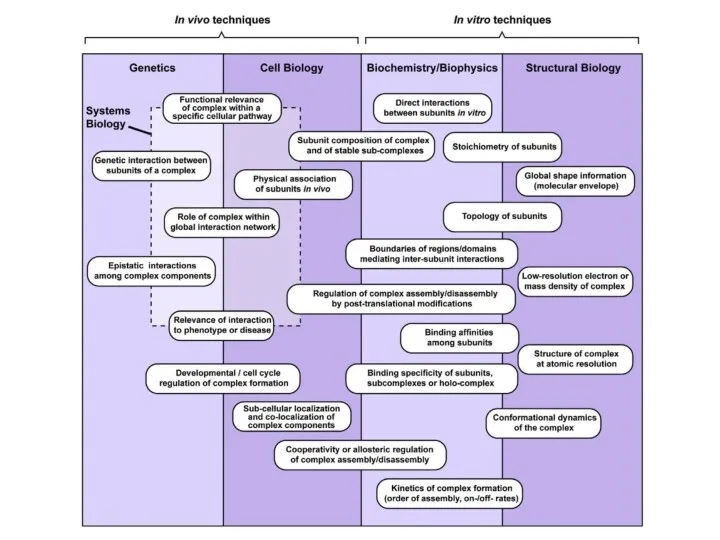
- 2. Protein-protein interactions (PPIs) >80% of proteins function via interaction with other proteins (PMID: 17640003) For each
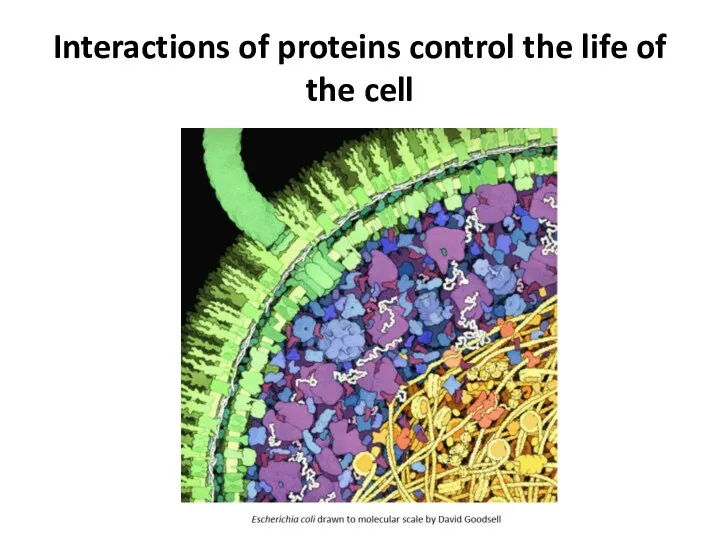
- 3. Interactions of proteins control the life of the cell
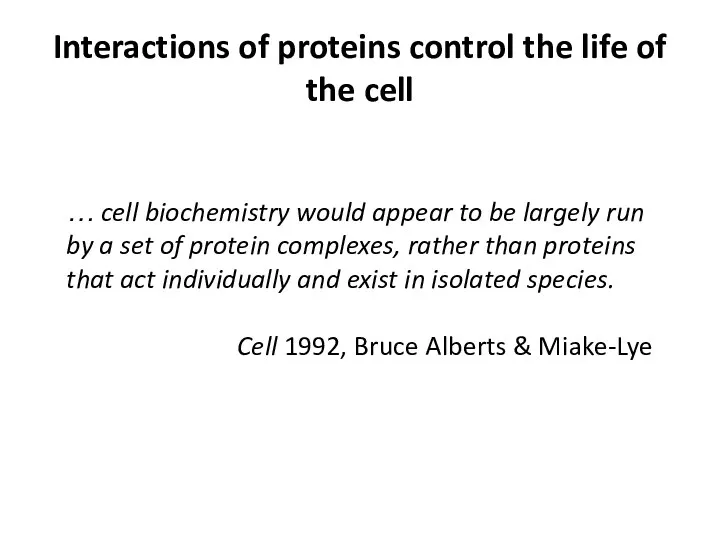
- 4. Interactions of proteins control the life of the cell … cell biochemistry would appear to be
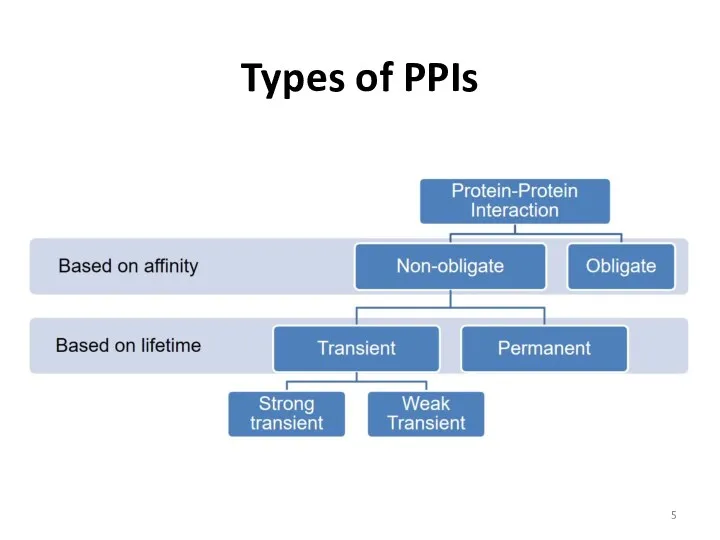
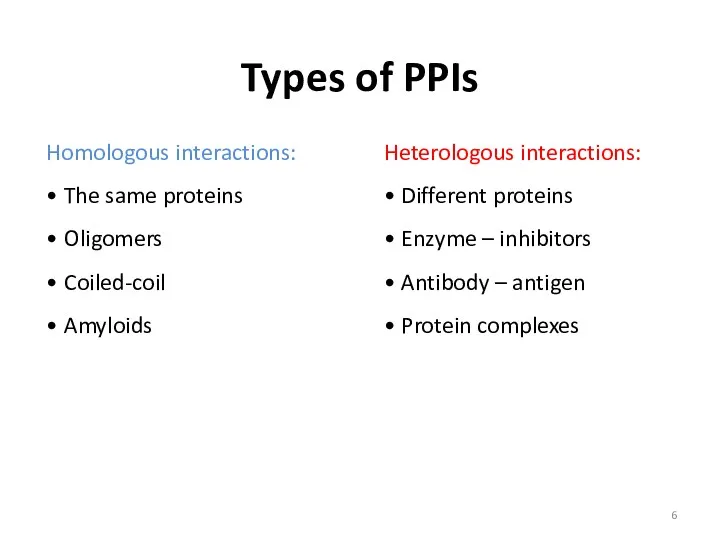
- 5. Types of PPIs
- 6. Types of PPIs Homologous interactions: • The same proteins • Oligomers • Coiled-coil • Amyloids Heterologous
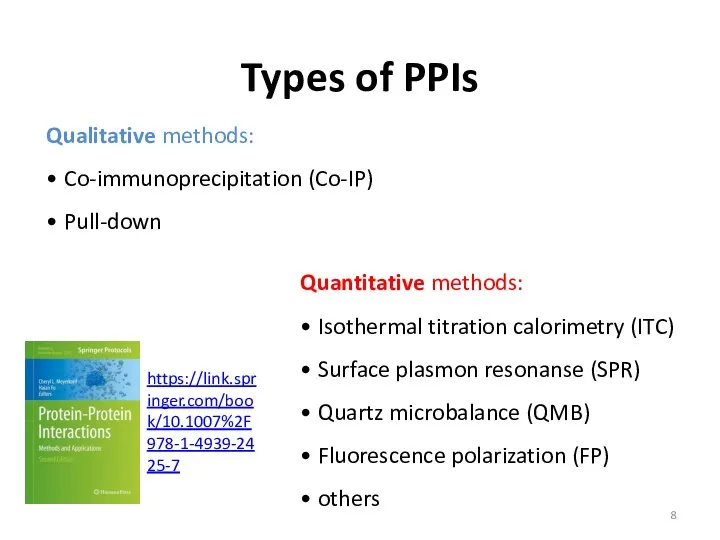
- 8. Types of PPIs Qualitative methods: • Co-immunoprecipitation (Co-IP) • Pull-down Quantitative methods: • Isothermal titration calorimetry
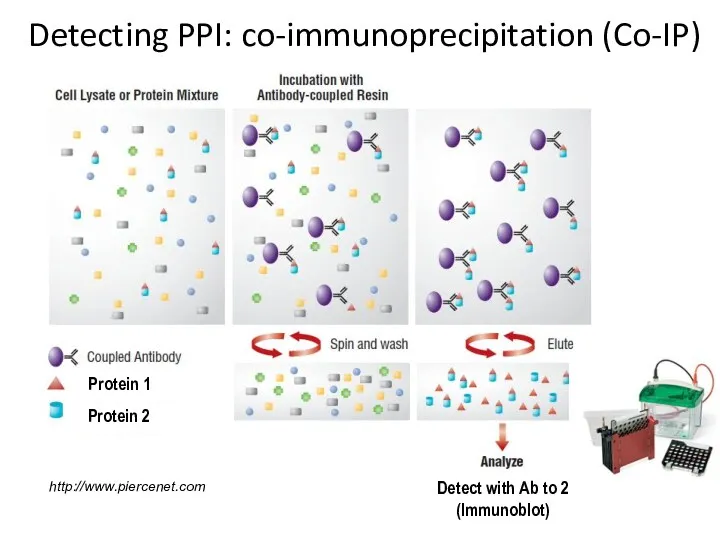
- 9. Detecting PPI: co-immunoprecipitation (Co-IP)
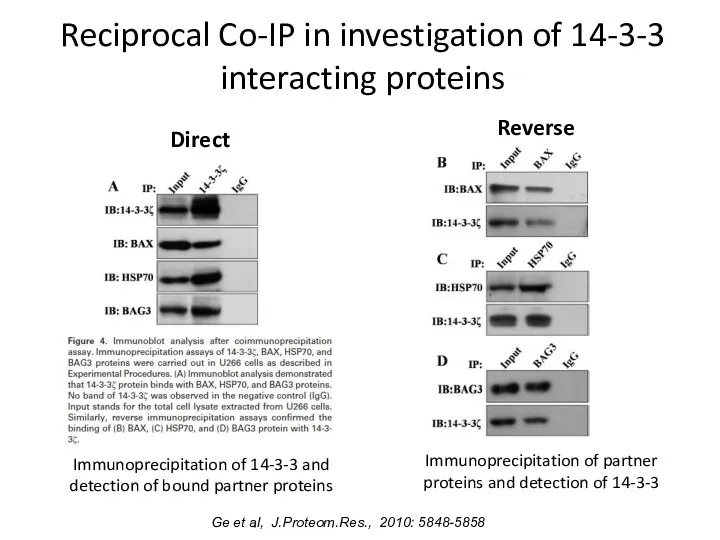
- 10. Reciprocal Co-IP in investigation of 14-3-3 interacting proteins Direct Immunoprecipitation of 14-3-3 and detection of bound
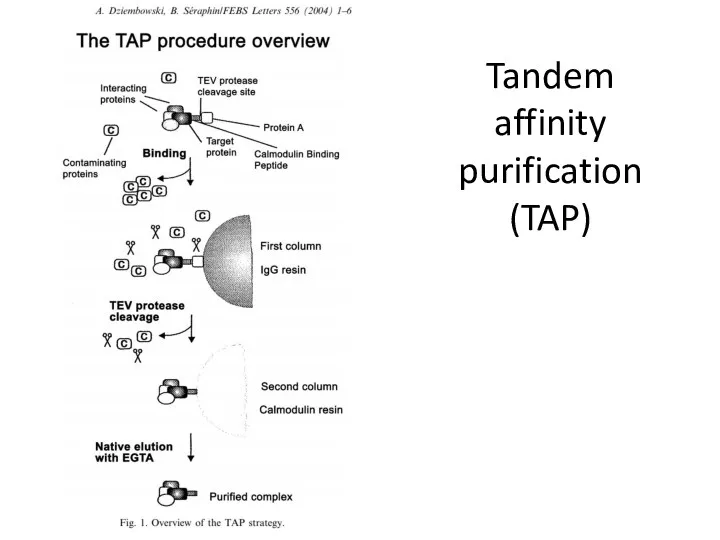
- 11. Tandem affinity purification (TAP)
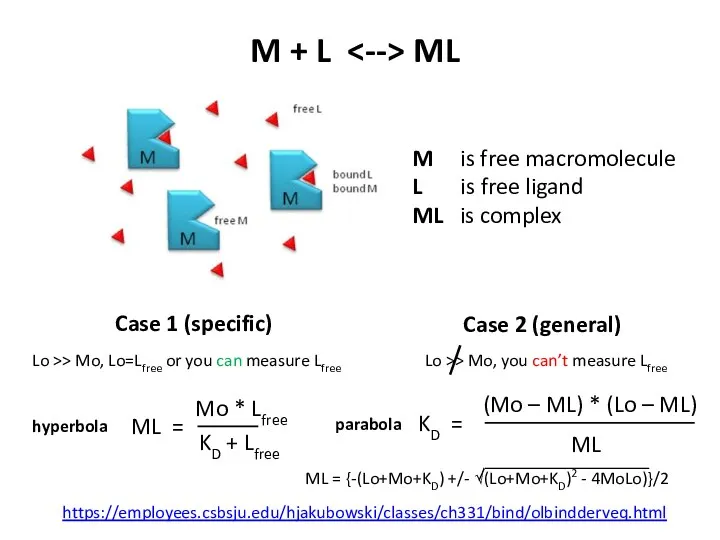
- 12. M + L ML M is free macromolecule L is free ligand ML is complex Lo
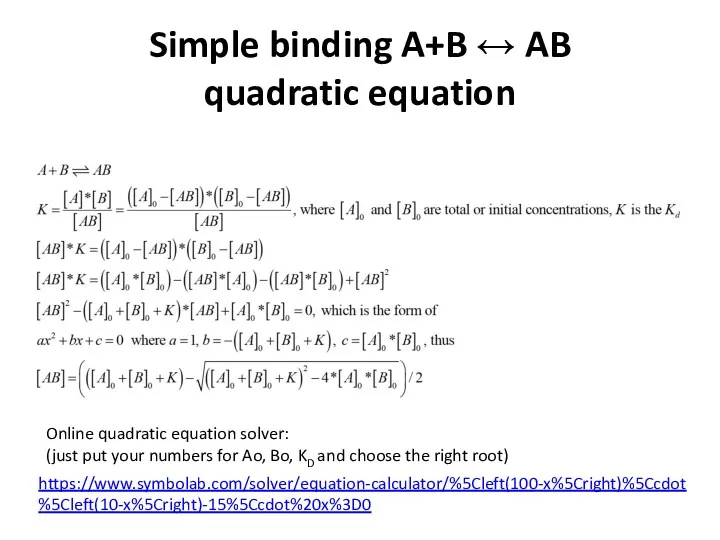
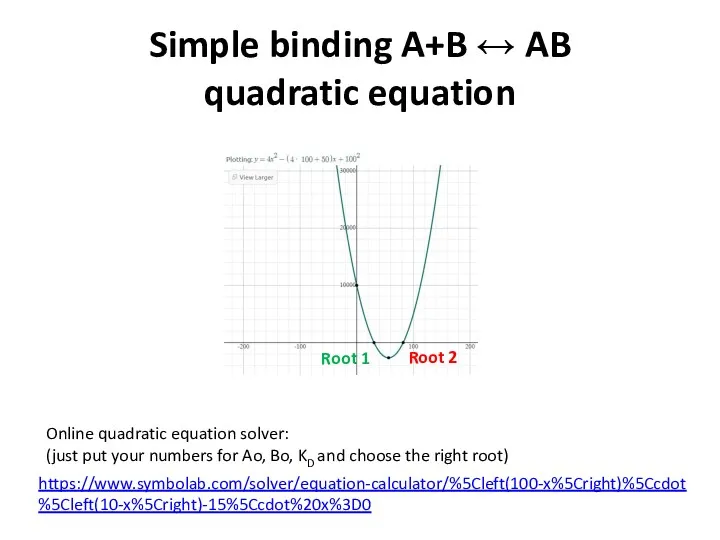
- 13. Simple binding A+B ↔ AB quadratic equation https://www.symbolab.com/solver/equation-calculator/%5Cleft(100-x%5Cright)%5Ccdot%5Cleft(10-x%5Cright)-15%5Ccdot%20x%3D0 Online quadratic equation solver: (just put your numbers
- 14. Simple binding A+B ↔ AB quadratic equation https://www.symbolab.com/solver/equation-calculator/%5Cleft(100-x%5Cright)%5Ccdot%5Cleft(10-x%5Cright)-15%5Ccdot%20x%3D0 Online quadratic equation solver: (just put your numbers
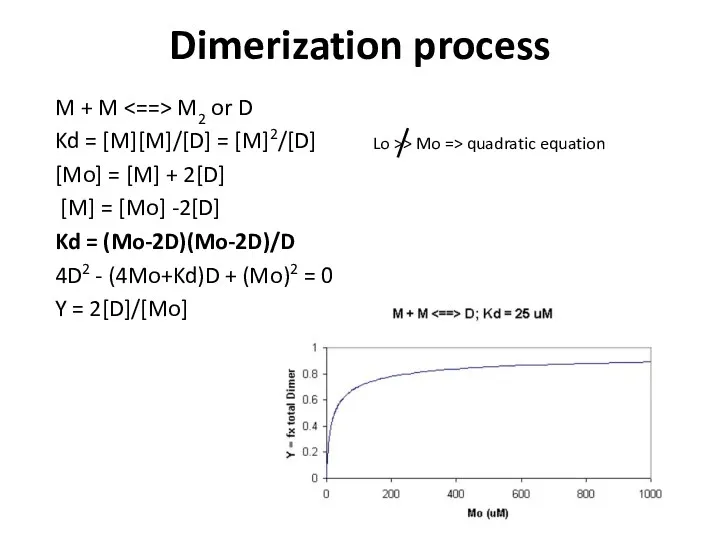
- 15. Dimerization process M + M M2 or D Kd = [M][M]/[D] = [M]2/[D] [Mo] = [M]
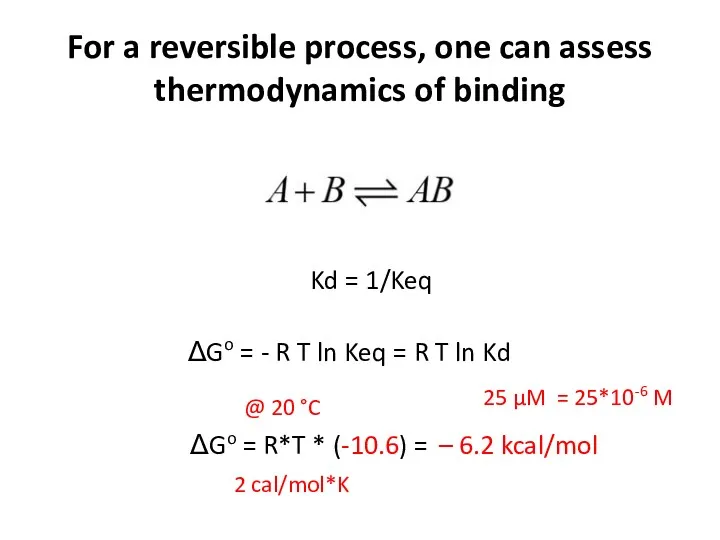
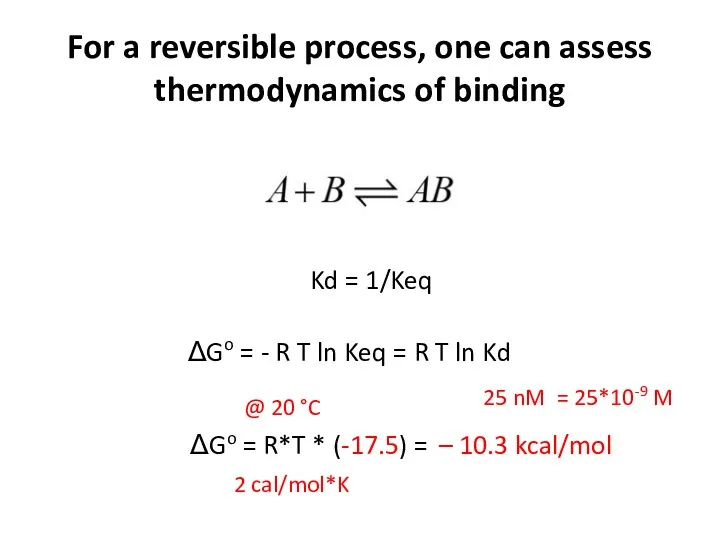
- 16. For a reversible process, one can assess thermodynamics of binding Kd = 1/Keq ΔGo = -
- 17. For a reversible process, one can assess thermodynamics of binding Kd = 1/Keq ΔGo = -
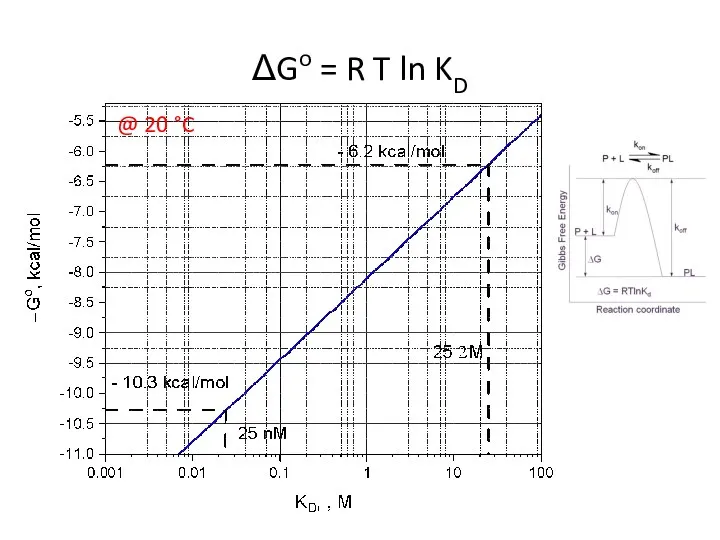
- 18. ΔGo = R T ln KD @ 20 °C
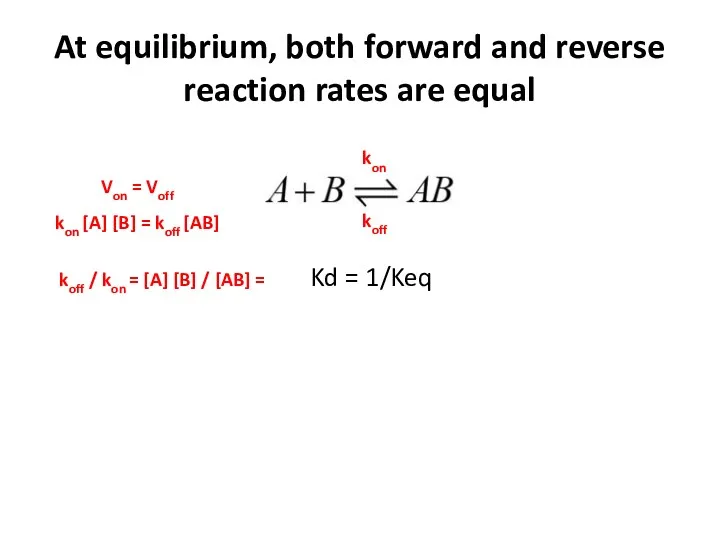
- 19. At equilibrium, both forward and reverse reaction rates are equal Kd = 1/Keq kon koff Von
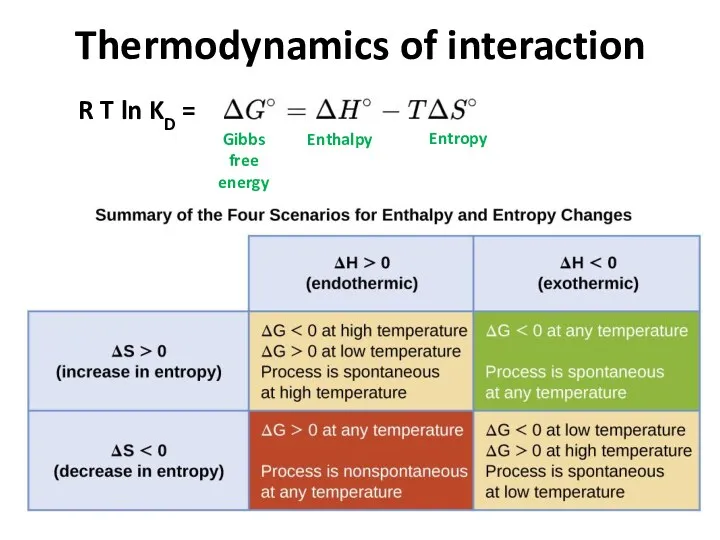
- 20. Thermodynamics of interaction Gibbs free energy Enthalpy Entropy R T ln KD =
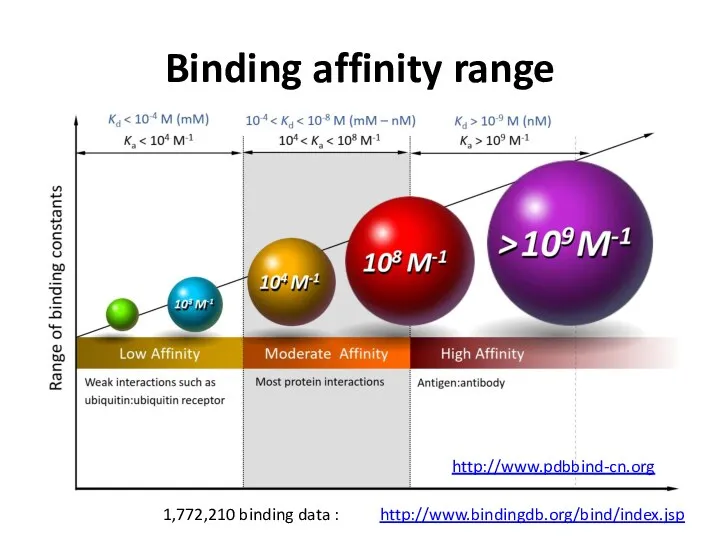
- 21. Binding affinity range http://www.bindingdb.org/bind/index.jsp 1,772,210 binding data : http://www.pdbbind-cn.org
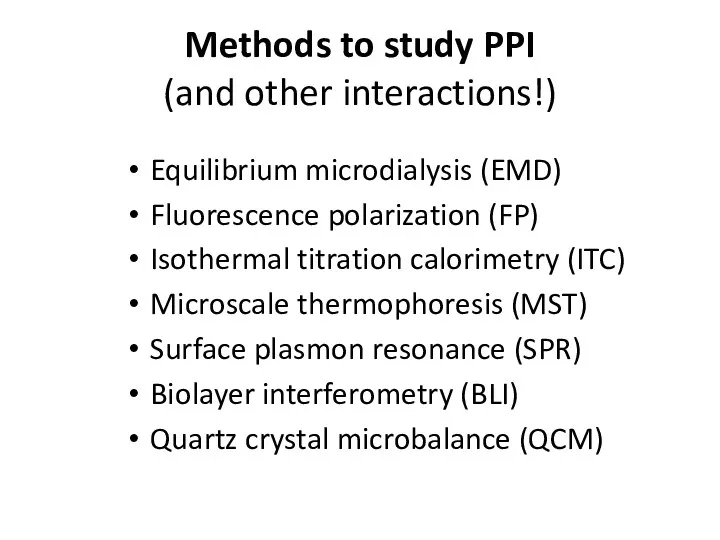
- 22. Methods to study PPI (and other interactions!) Equilibrium microdialysis (EMD) Fluorescence polarization (FP) Isothermal titration calorimetry
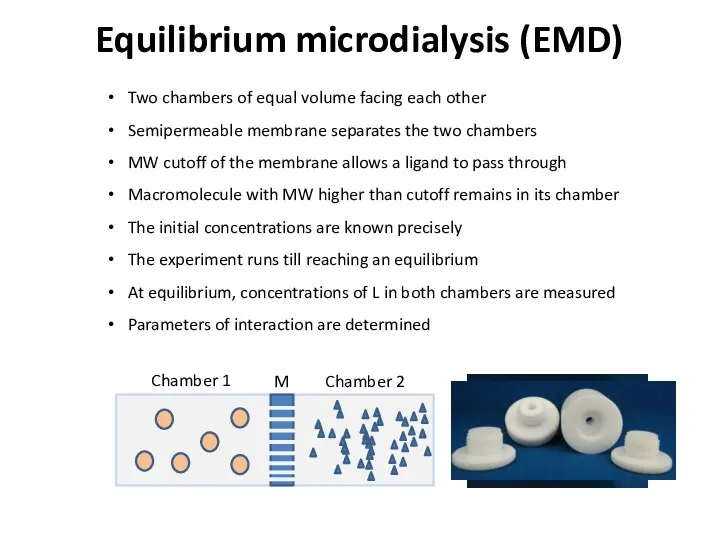
- 23. Equilibrium microdialysis (EMD) Two chambers of equal volume facing each other Semipermeable membrane separates the two
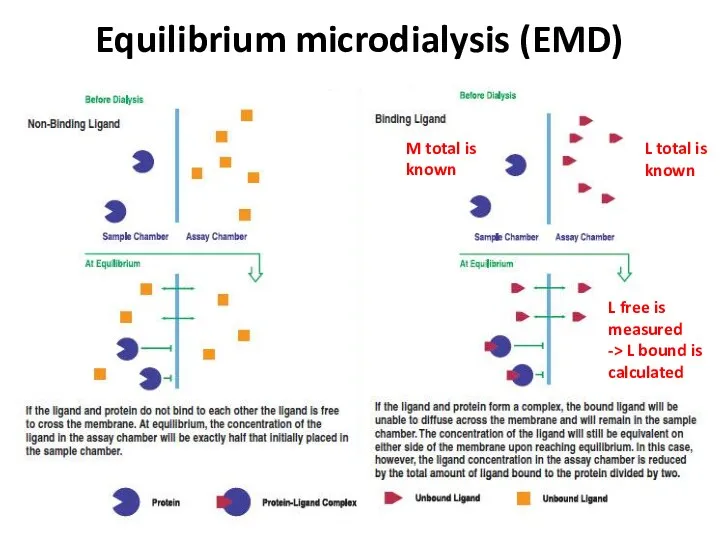
- 24. Equilibrium microdialysis (EMD) L total is known L free is measured -> L bound is calculated
- 25. Equilibrium microdialysis (EMD) KD = [M] * [L] [ML] M + L ML Fast Easy Inexpensive
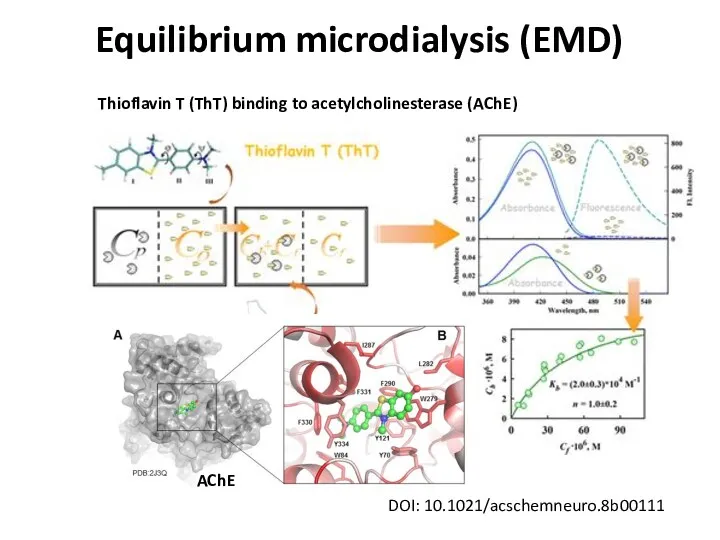
- 26. Equilibrium microdialysis (EMD) DOI: 10.1021/acschemneuro.8b00111 Thioflavin T (ThT) binding to acetylcholinesterase (AChE) AChE
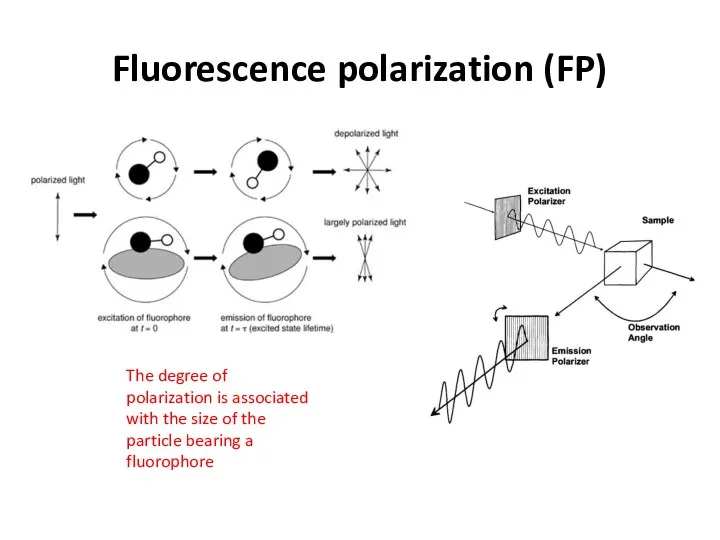
- 27. Fluorescence polarization (FP) The degree of polarization is associated with the size of the particle bearing
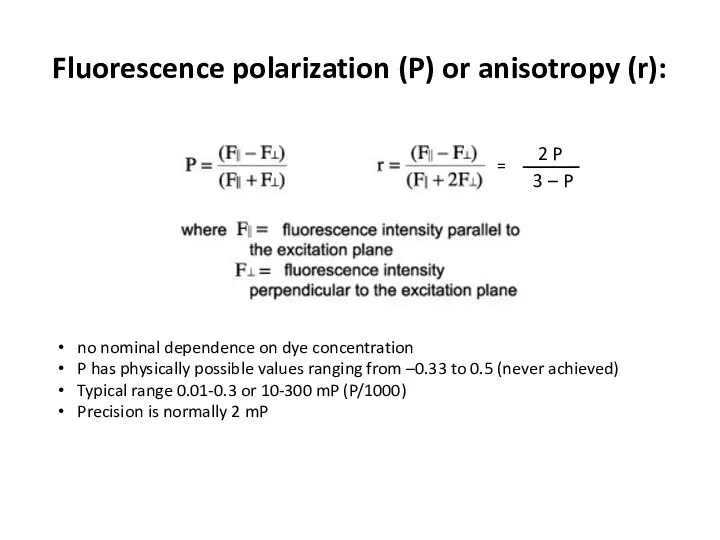
- 28. Fluorescence polarization (P) or anisotropy (r): no nominal dependence on dye concentration P has physically possible
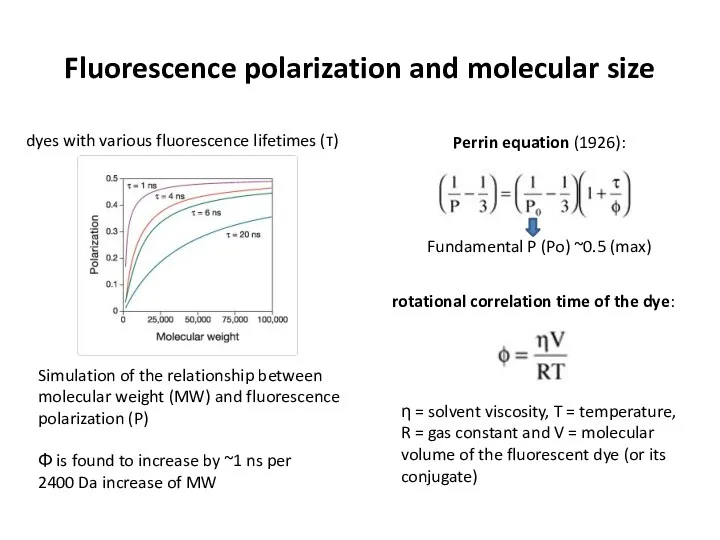
- 29. Fluorescence polarization and molecular size η = solvent viscosity, T = temperature, R = gas constant
- 30. FP features Great tool to study interactions Small sample consumption Low limit of detection Rapid response
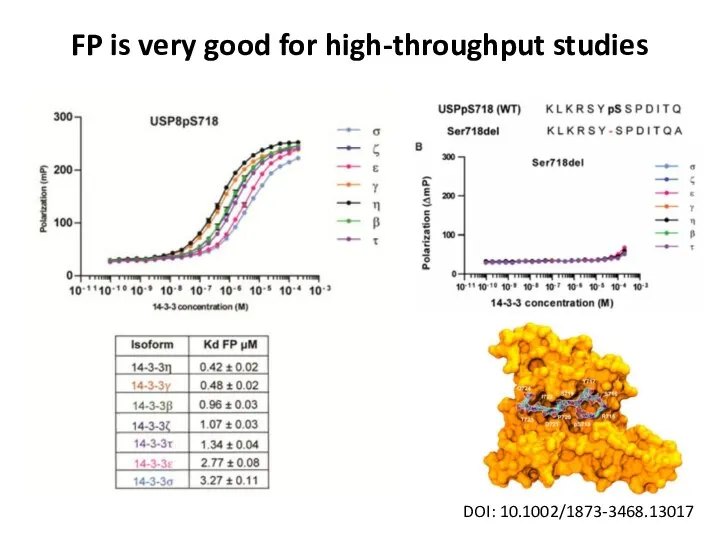
- 31. FP is very good for high-throughput studies DOI: 10.1002/1873-3468.13017
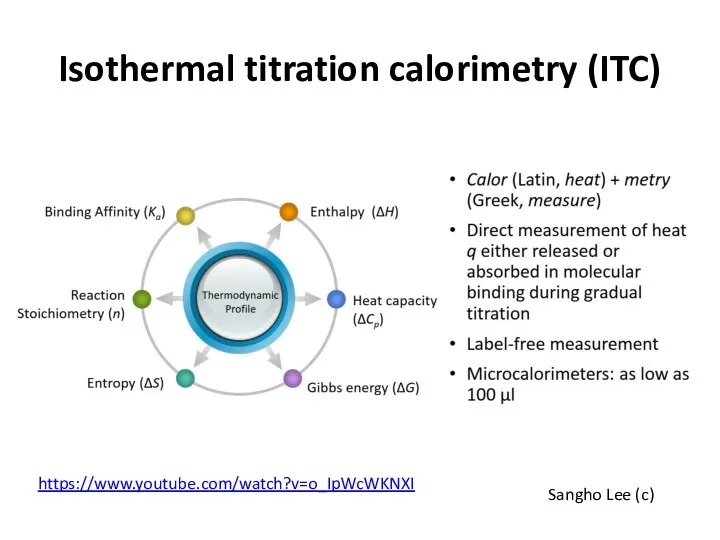
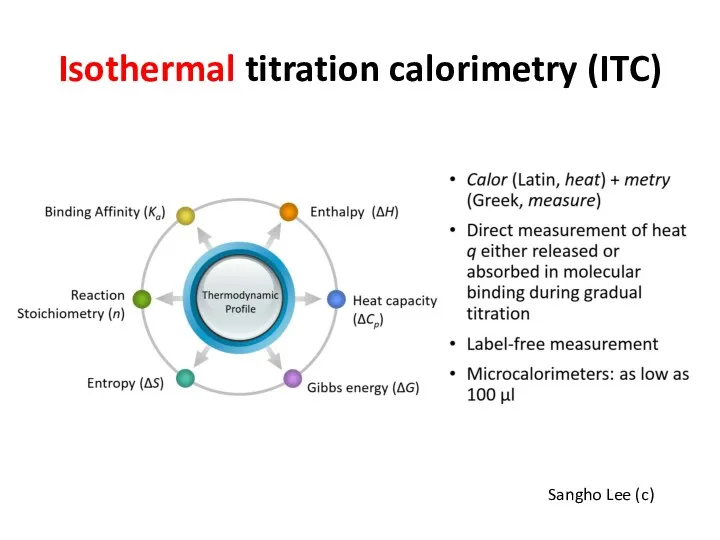
- 32. Isothermal titration calorimetry (ITC) Sangho Lee (c) https://www.youtube.com/watch?v=o_IpWcWKNXI
- 33. Isothermal titration calorimetry (ITC) Sangho Lee (c)
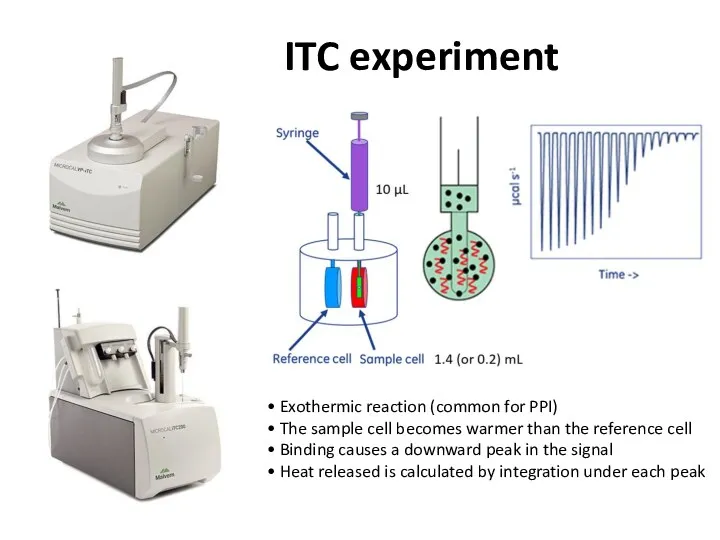
- 34. ITC experiment • Exothermic reaction (common for PPI) • The sample cell becomes warmer than the
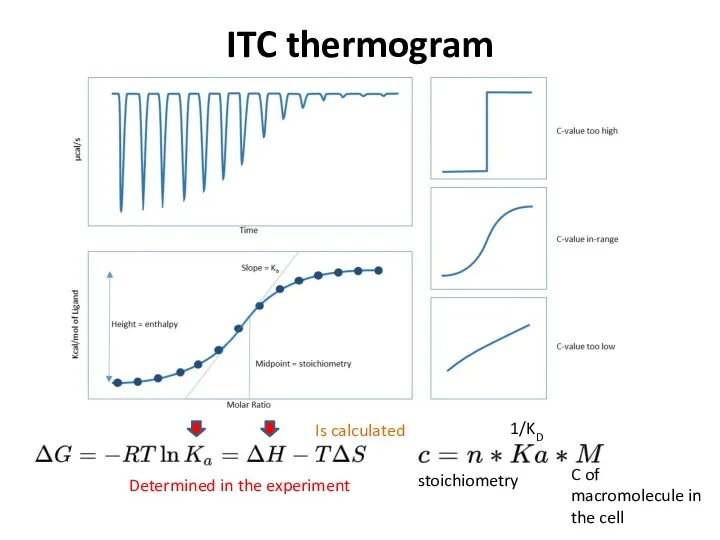
- 35. ITC thermogram stoichiometry 1/KD C of macromolecule in the cell Determined in the experiment Is calculated
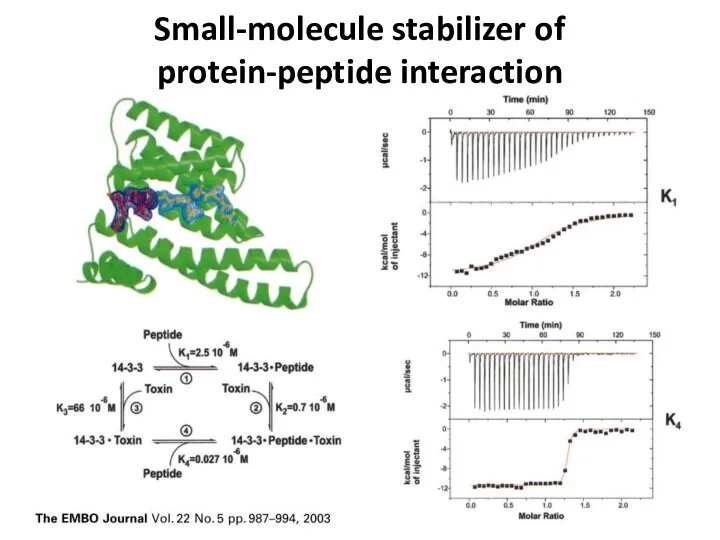
- 36. Small-molecule stabilizer of protein-peptide interaction
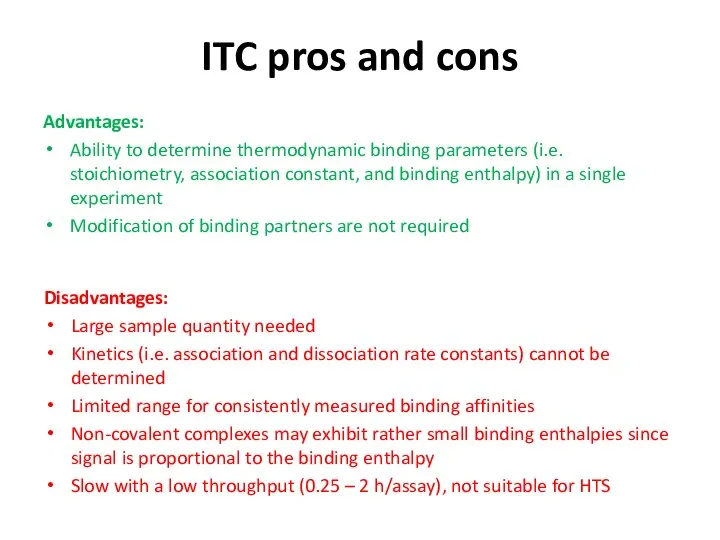
- 37. ITC pros and cons Advantages: Ability to determine thermodynamic binding parameters (i.e. stoichiometry, association constant, and
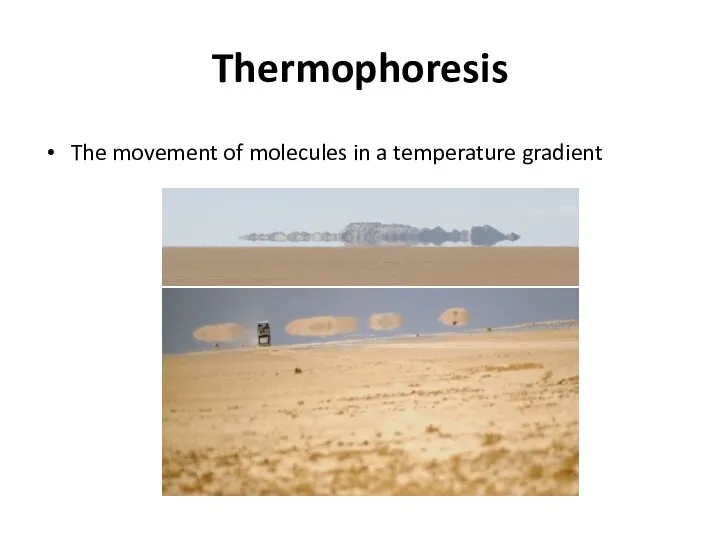
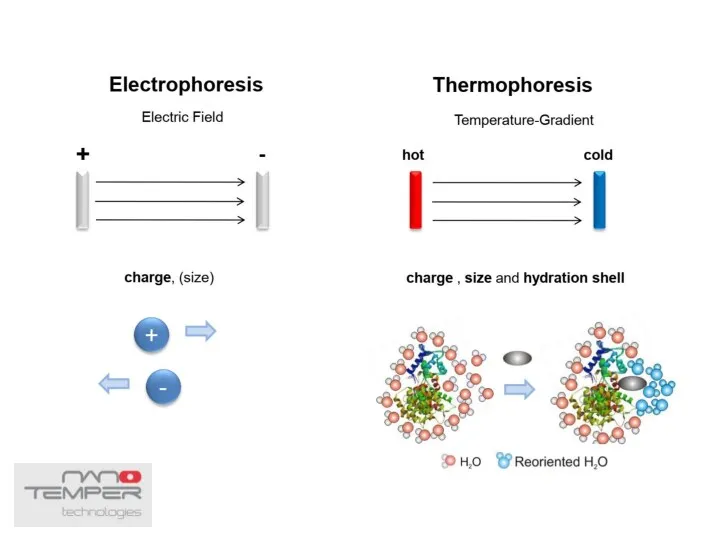
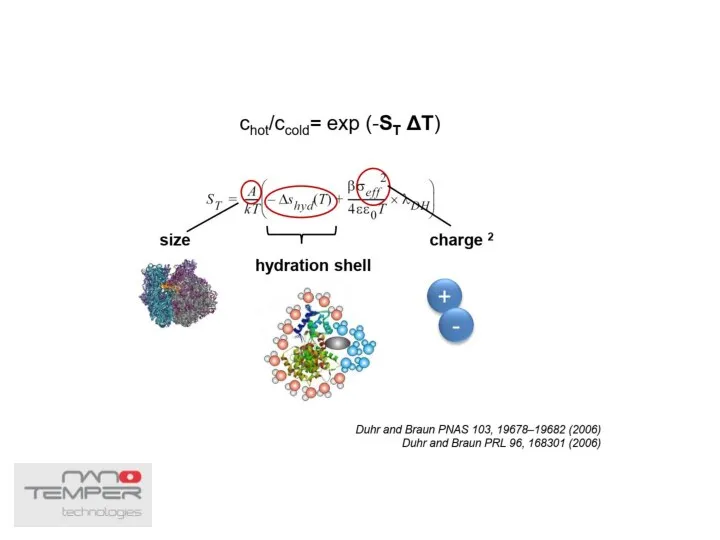
- 38. Thermophoresis The movement of molecules in a temperature gradient
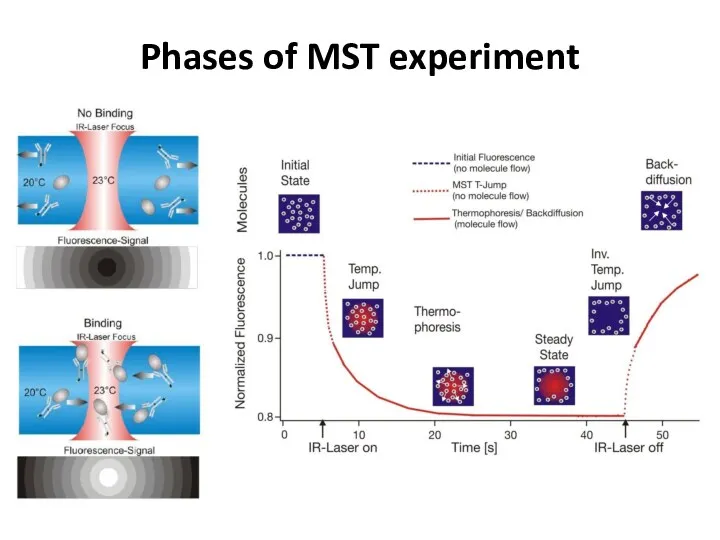
- 41. Phases of MST experiment
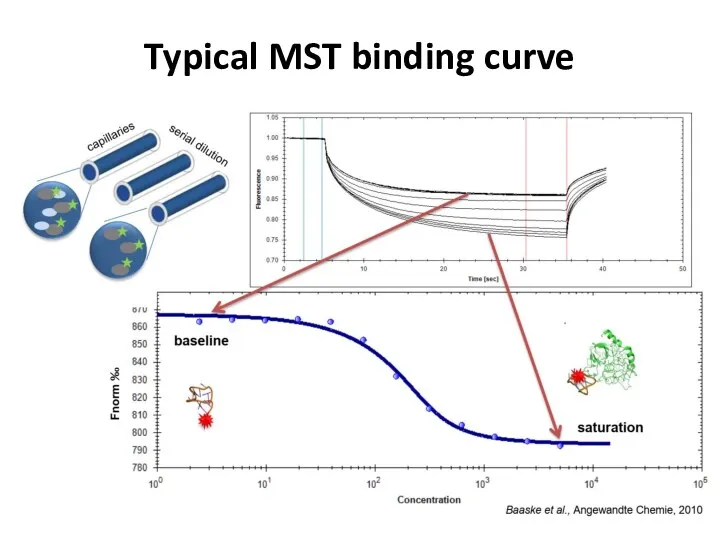
- 42. Typical MST binding curve
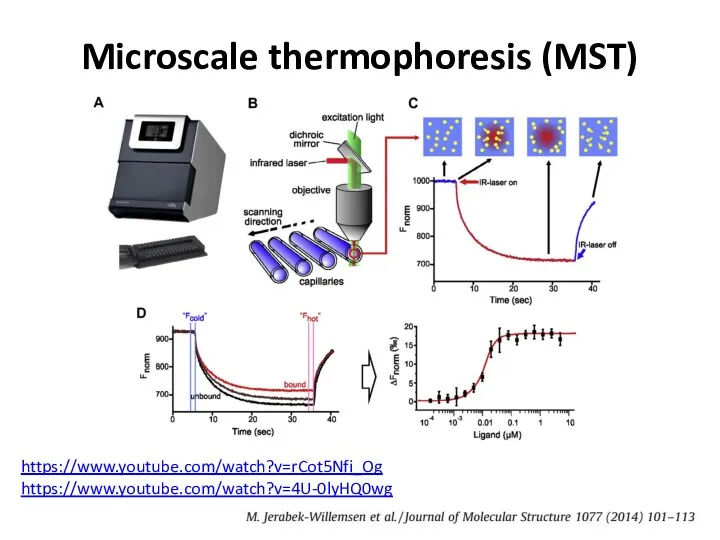
- 43. Microscale thermophoresis (MST) https://www.youtube.com/watch?v=4U-0lyHQ0wg https://www.youtube.com/watch?v=rCot5Nfi_Og
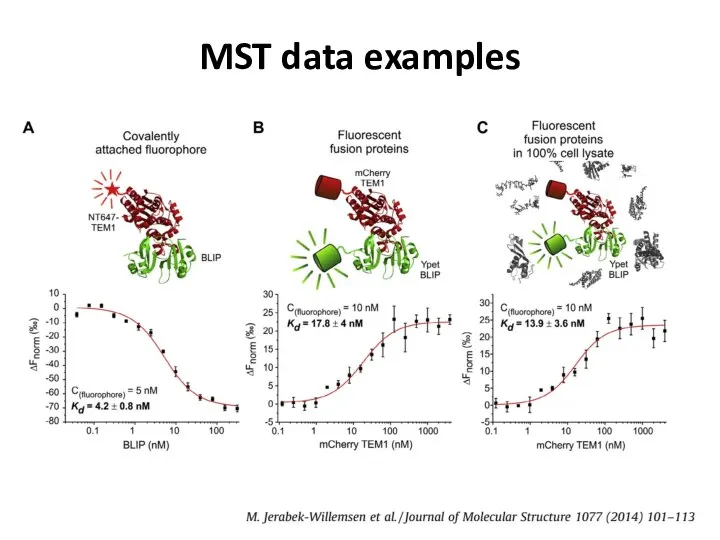
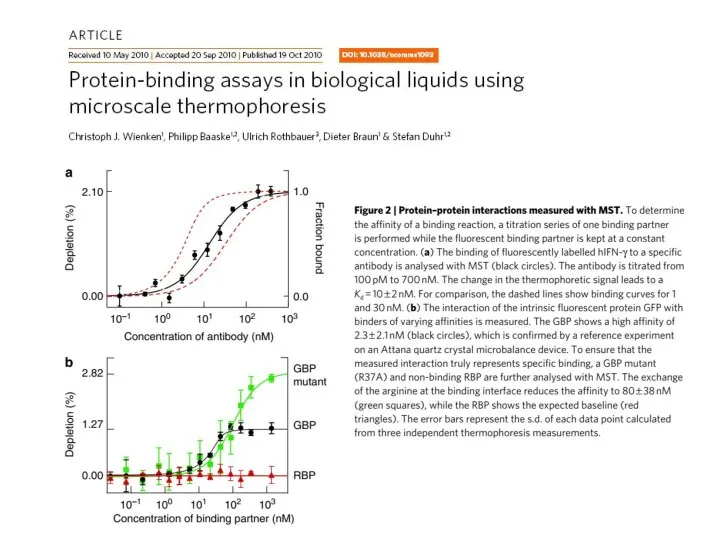
- 44. MST data examples
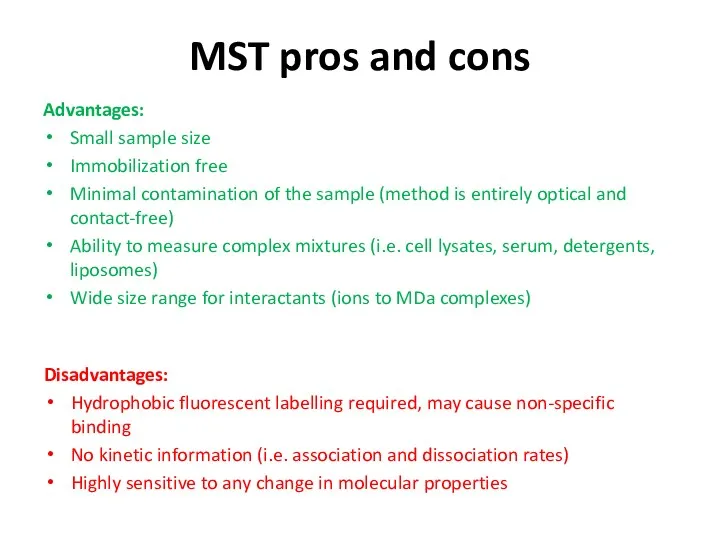
- 46. MST pros and cons Advantages: Small sample size Immobilization free Minimal contamination of the sample (method
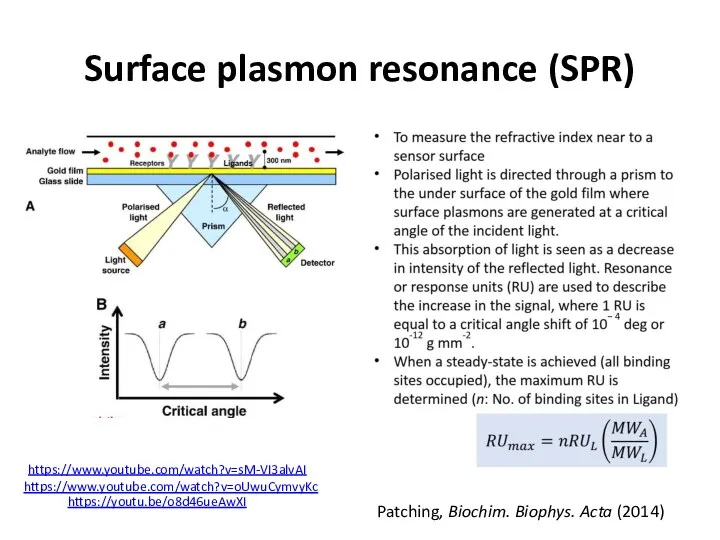
- 47. Surface plasmon resonance (SPR)
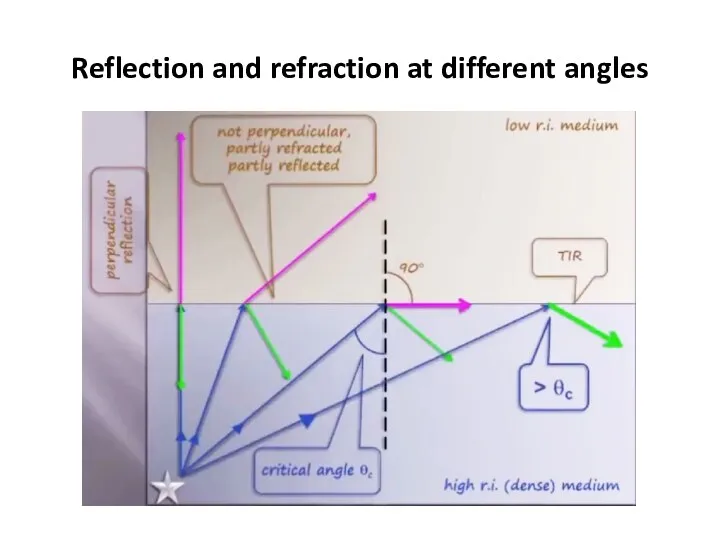
- 48. Reflection and refraction at different angles
- 49. Surface plasmon resonance (SPR) Patching, Biochim. Biophys. Acta (2014) https://youtu.be/o8d46ueAwXI https://www.youtube.com/watch?v=oUwuCymvyKc https://www.youtube.com/watch?v=sM-VI3alvAI
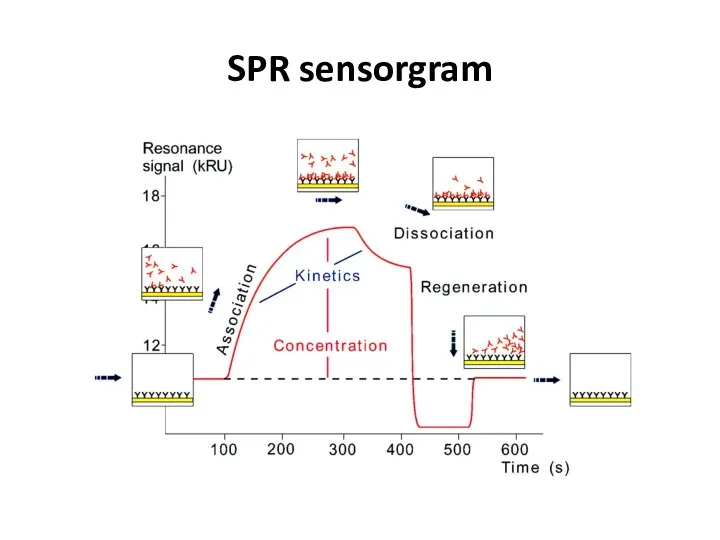
- 50. SPR sensorgram
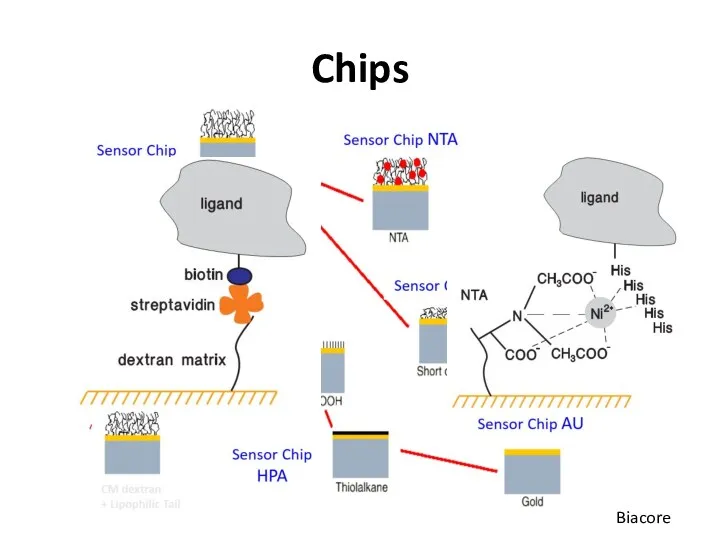
- 51. Chips Biacore
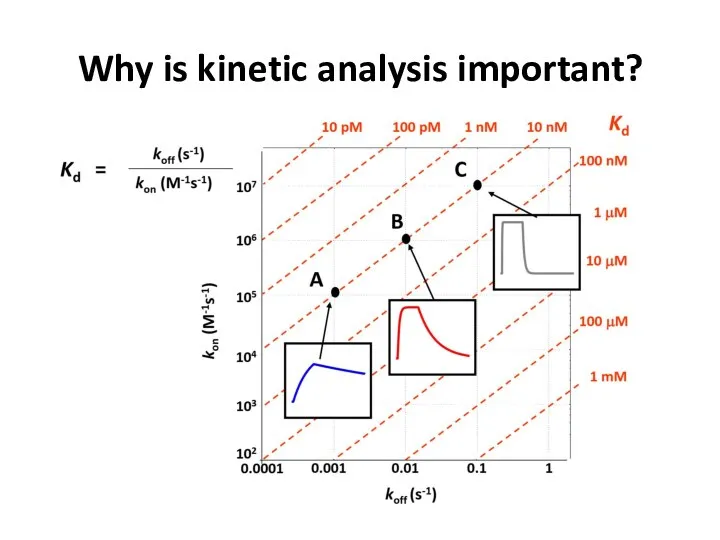
- 52. Why is kinetic analysis important?
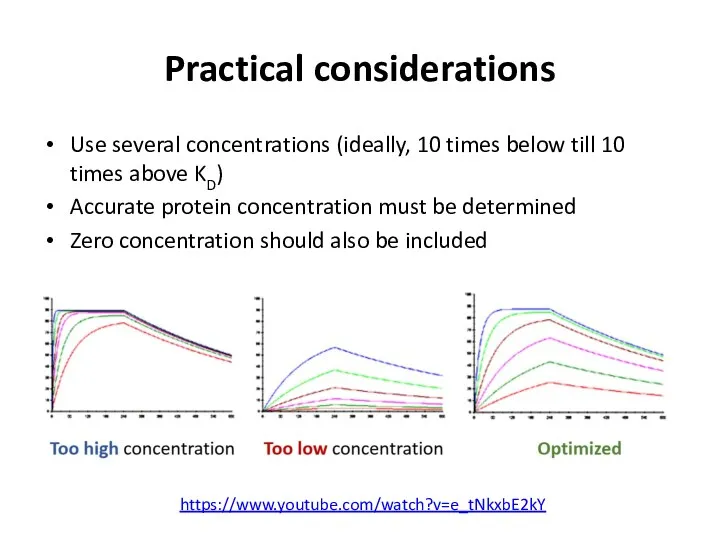
- 53. Practical considerations Use several concentrations (ideally, 10 times below till 10 times above KD) Accurate protein
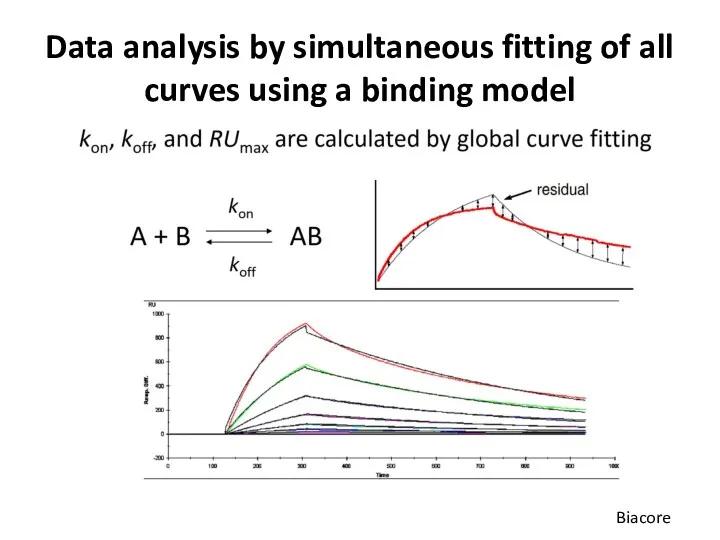
- 54. Data analysis by simultaneous fitting of all curves using a binding model Biacore
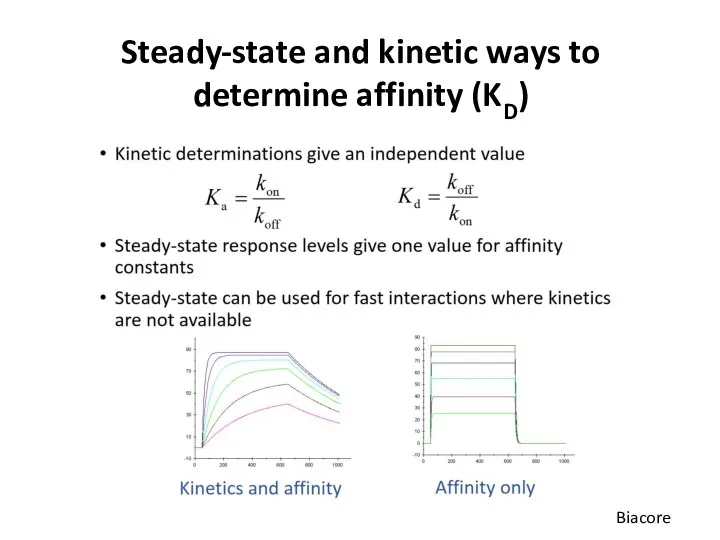
- 55. Steady-state and kinetic ways to determine affinity (KD) Biacore
- 56. Steady-state and kinetic ways to determine affinity (KD) Biacore
- 57. SPR pros and cons Advantages: Label-free detection Real-time data (i.e. quantitative binding affinities, kinetics and thermodynamics)
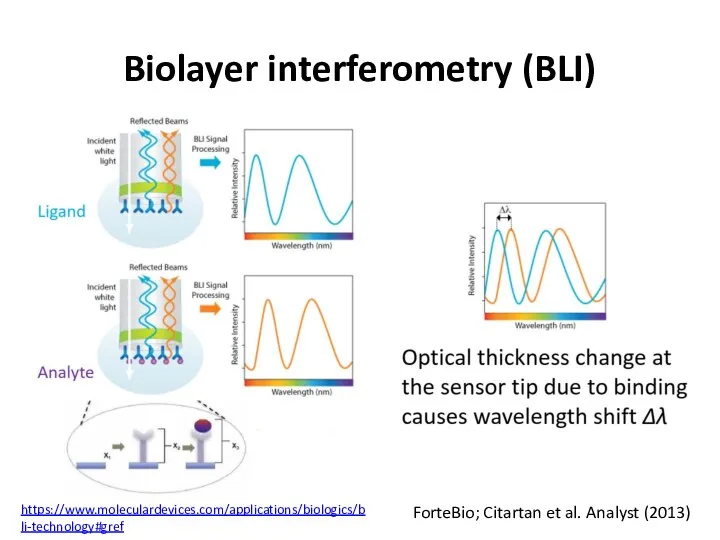
- 58. Biolayer interferometry (BLI) ForteBio; Citartan et al. Analyst (2013) https://www.moleculardevices.com/applications/biologics/bli-technology#gref
- 59. Instruments 8 channels 1 channel
- 60. Instruments 8 channels 1 channel
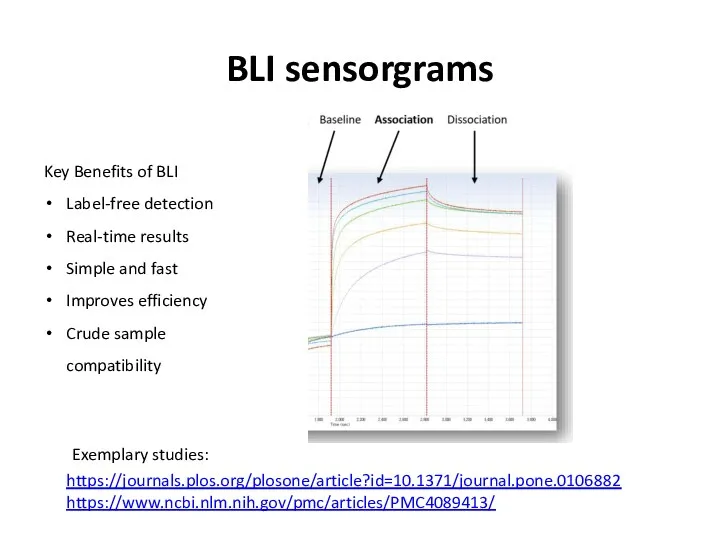
- 61. BLI sensorgrams Key Benefits of BLI Label-free detection Real-time results Simple and fast Improves efficiency Crude
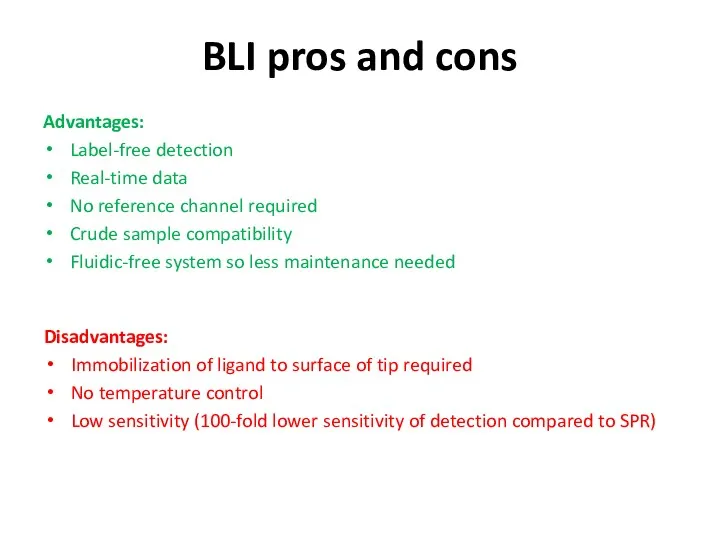
- 62. BLI pros and cons Advantages: Label-free detection Real-time data No reference channel required Crude sample compatibility
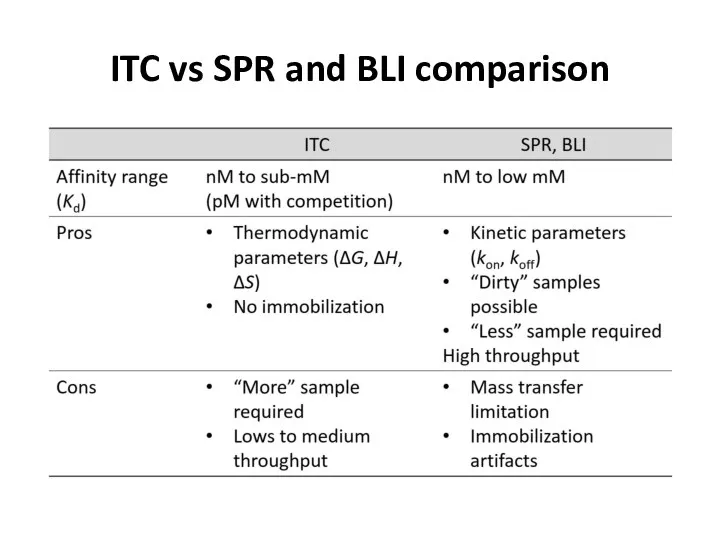
- 63. ITC vs SPR and BLI comparison
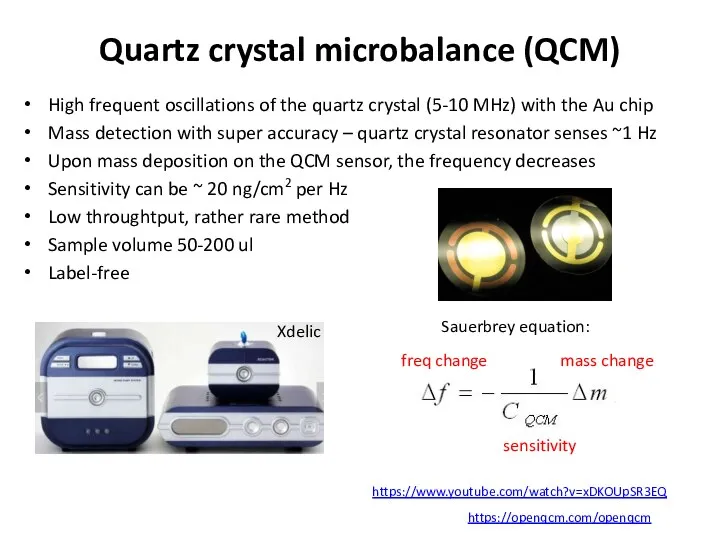
- 64. Quartz crystal microbalance (QCM) High frequent oscillations of the quartz crystal (5-10 MHz) with the Au
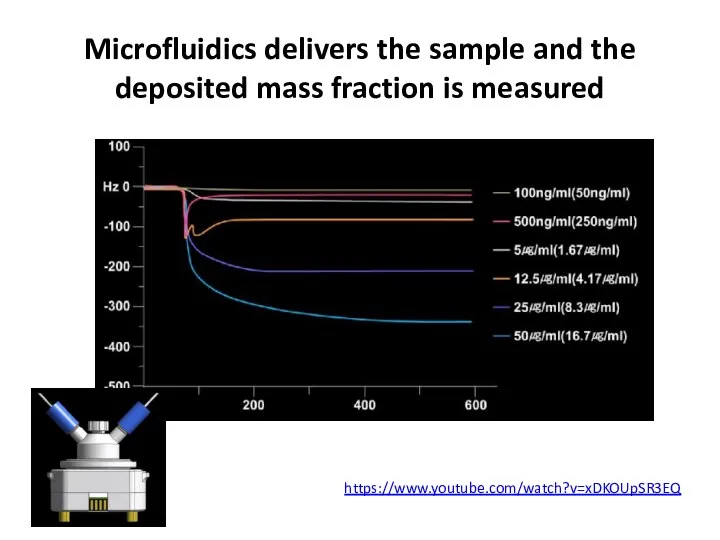
- 65. Microfluidics delivers the sample and the deposited mass fraction is measured https://www.youtube.com/watch?v=xDKOUpSR3EQ
- 67. Скачать презентацию
![Equilibrium microdialysis (EMD) KD = [M] * [L] [ML] M](/_ipx/f_webp&q_80&fit_contain&s_1440x1080/imagesDir/jpg/400849/slide-24.jpg)






 Кристаллические решётки и их виды
Кристаллические решётки и их виды Enantioselective Total Synthesis
Enantioselective Total Synthesis Химические свойства солей
Химические свойства солей Закон постоянства состава. Молекулярная формула вещества
Закон постоянства состава. Молекулярная формула вещества Группа ультраосновных пород
Группа ультраосновных пород Строение атома
Строение атома Серная кислота
Серная кислота 20230419_izomery
20230419_izomery Бор шикізатын қышқылдық ыдырату
Бор шикізатын қышқылдық ыдырату kremniy
kremniy Алкены. Непредельные углеводороды
Алкены. Непредельные углеводороды Типы химических реакций. Систематизация и обобщение знаний
Типы химических реакций. Систематизация и обобщение знаний Висмут, ртуть, сурьма
Висмут, ртуть, сурьма Метаболизм нуклеиновых кислот
Метаболизм нуклеиновых кислот Методы получения органических галогенидов
Методы получения органических галогенидов Неметали. Фізичні та хімічні властивості. Явище адсорбції. Сполуки неметалічних елементів з Гідрогеном
Неметали. Фізичні та хімічні властивості. Явище адсорбції. Сполуки неметалічних елементів з Гідрогеном Химия аминокислот. Лекция № 4
Химия аминокислот. Лекция № 4 Стратегия химической промышленности
Стратегия химической промышленности Вещества
Вещества Химия 20 века
Химия 20 века Газовые смеси
Газовые смеси Правила техники безопасности при работе в химическом кабинете
Правила техники безопасности при работе в химическом кабинете Формы парфюмерно-косметической продукции
Формы парфюмерно-косметической продукции Химические свойства основных неорганических соединений в свете ЭД и ОВР. 9 класс
Химические свойства основных неорганических соединений в свете ЭД и ОВР. 9 класс Создание косметических средств
Создание косметических средств Аллотропия. Аллотропные модификации
Аллотропия. Аллотропные модификации Электрохимический ряд напряжений металлов
Электрохимический ряд напряжений металлов Металлы в природе. Способы получения металлов
Металлы в природе. Способы получения металлов